18.6 The Evolutionary History of a Group of Organisms Can Be Reconstructed by Studying Changes in Homologous Characteristics
The evolutionary relationships among a group of organisms are termed a phylogeny. Because most evolution takes place over long periods and is not amenable to direct observation, biologists must reconstruct phylogenies by inferring the evolutionary relationships among present-day organisms. The discovery of fossils of ancestral organisms can aid in the reconstruction of phylogenies, but the fossil record is often too poor to be of much help. Thus, biologists are often restricted to the analysis of characteristics in present-day organisms to determine their evolutionary relationships. In the past, phylogenetic relationships were reconstructed on the basis of phenotypic characteristics—often, anatomical traits. Today, molecular data, including protein and DNA sequences, are frequently used to construct phylogenetic trees.
Phylogenies are reconstructed by inferring changes that have taken place in homologous characteristics: characteristics that have evolved from the same character in a common ancestor. For example, although the front leg of a mouse and the wing of a bat look different and have different functions, close examination of their structure and development reveals that they are indeed homologous; both evolved from the forelimb of an early mammal that was an ancestor to both mouse and bat. And, because mouse and bat have these homologous features and others in common, we know that they are both mammals. Similarly, DNA sequences are homologous if two present-day sequences evolved from a single sequence found in an ancestor. For example, all eukaryotic organisms have a gene for cytochrome c, an enzyme that helps carry out oxidative respiration. This gene is assumed to have arisen in a single organism in the distant past and was then passed down to descendants of that early ancestor. Today, all copies of the gene for cytochrome c are homologous because they all evolved from the same original copy in the distant ancestor of all organisms that possess this gene.
Page 489
A graphical representation of a phylogeny is called a phylogenetic tree. As shown in Figure 18.15, a phylogenetic tree depicts the evolutionary relationships among different organisms, similarly to the way in which a pedigree represents the genealogical relationships among family members. A phylogenetic tree consists of nodes that represent the different organisms being compared, which might be different individuals, populations, or species. Terminal nodes (those at the ends of the outermost branches of the tree) represent organisms for which data have been obtained, usually present-day organisms. Internal nodes represent common ancestors that existed before divergence between organisms took place. In most cases, the internal nodes represent past ancestors that are inferred from the analysis. The nodes are connected by branches, which represent the evolutionary connections between organisms. In some phylogenetic trees, the lengths of the branches represent the amount of evolutionary divergence that has taken place between organisms. When one internal node represents a common ancestor to all other nodes on the tree, the tree is said to be rooted. Trees are often rooted by including in the analysis a present-day organism that is related to the group, but more distantly than the other members of the phylogeny; this distantly related organism is referred to as an outgroup.
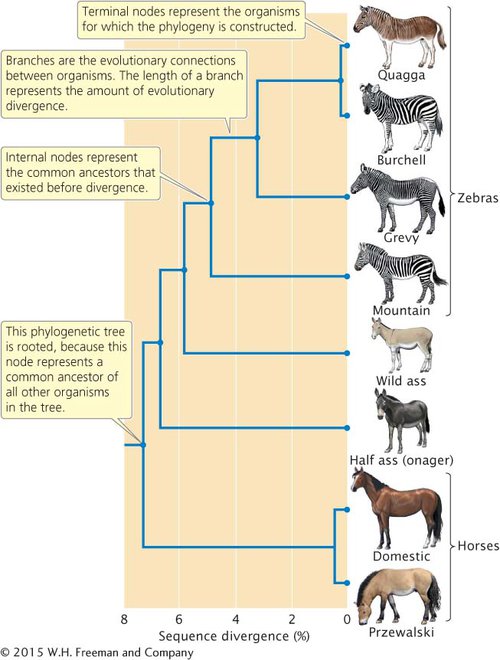
18.15 A phylogenetic tree is a graphical representation of the evolutionary relationships among a group of organisms.
Page 490
CONCEPTS
A phylogeny represents the evolutionary relationships among a group of organisms. It is often depicted graphically by a phylogenetic tree, which consists of nodes representing the organisms and branches representing their evolutionary connections.
