Effects of Multiple Crossovers
So far, we have examined the effects of double crossovers that take place between only two of the four chromatids (strands) of a homologous pair. These crossovers are called two-strand crossovers. Double crossovers that include three and even four of the chromatids of a homologous pair may also take place (Figure 5.15). If we examine only the alleles at loci on either side of both crossover events, two-strand double crossovers result in no new combinations of alleles, and no recombinant gametes are produced (see Figure 5.15). Three-strand double crossovers result in two of the four gametes being recombinant, and four-strand double crossovers result in all four gametes being recombinant. Thus, two-strand double crossovers produce 0% recombination, three-strand double crossovers produce 50% recombination, and four-strand double crossovers produce 100% recombination. The overall result is that all types of double crossovers, taken together, produce an average of 50% recombinant progeny.
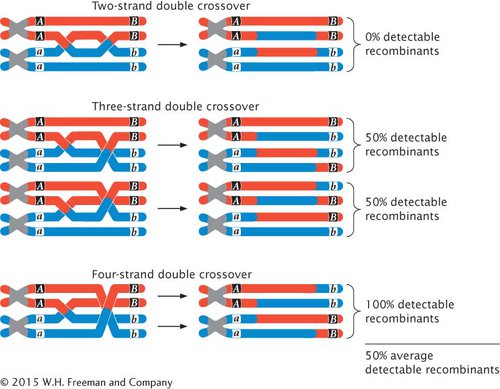
5.15 Results of two-, three-, and four-strand double crossovers on recombination between two genes.
Page 138
As we have seen, two-strand double crossovers cause alleles on either side of the crossovers to remain the same and produce no recombinant progeny. Three-strand and four-strand crossovers produce recombinant progeny, but these progeny are the same types as those produced by single crossovers. Consequently, some multiple crossovers go undetected when the progeny of a genetic cross are observed. Therefore, map distances based on recombination rates will underestimate the true physical distances between genes because some multiple crossovers will not be detected among the progeny of a cross. When genes are very close together, multiple crossovers are unlikely, and genetic distances based on recombination rates accurately correspond to the physical distances on the chromosome. But as the distance between genes increases, more multiple crossovers are likely, and the discrepancy between genetic distances (based on recombination rates) and physical distances increases. To correct for this discrepancy, geneticists have developed mathematical mapping functions, which relate recombination frequencies to actual physical distances between genes (Figure 5.16). Most of these functions are based on the Poisson distribution, which predicts the probability of multiple rare events. With the use of such mapping functions, map distances based on recombination rates can be more accurately estimated.
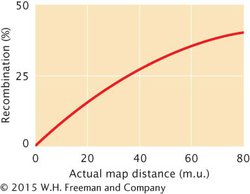
5.16 Recombination frequency underestimates the true physical distance between genes at higher map distances.



