Duplications
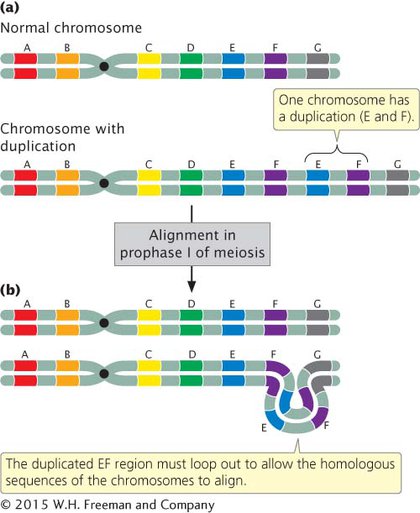
A chromosome duplication is a mutation in which part of the chromosome has been doubled (see Figure 6.4a). Consider a chromosome with segments AB•CDEFG, in which • represents the centromere. A duplication might include the EF segments, giving rise to a chromosome with segments AB•CDEFEFG. This type of duplication, in which the duplicated segment is immediately adjacent to the original segment, is called a tandem duplication. If the duplicated segment is located some distance from the original segment, either on the same chromosome or on a different one, the chromosome rearrangement is called a displaced duplication. An example of a displaced duplication would be AB•CDEFGEF. A duplication can be either in the same orientation as that of the original sequence, as in the two preceding examples, or inverted: AB•CDEFFEG. When the duplication is inverted, it is called a reverse duplication.
EFFECTS OF CHROMOSOME DUPLICATIONS An individual that is homozygous for a duplication carries that duplication on both homologous chromosomes, and an individual that is heterozygous for a duplication has one normal chromosome and one chromosome with the duplication. In heterozygotes (Figure 6.5a), problems in chromosome pairing arise at prophase I of meiosis because the two chromosomes are not homologous throughout their length. The pairing and synapsis of homologous regions require that one or both chromosomes loop and twist so that these regions are able to line up (Figure 6.5b). The appearance of this characteristic loop structure in meiosis is one way to detect duplications.
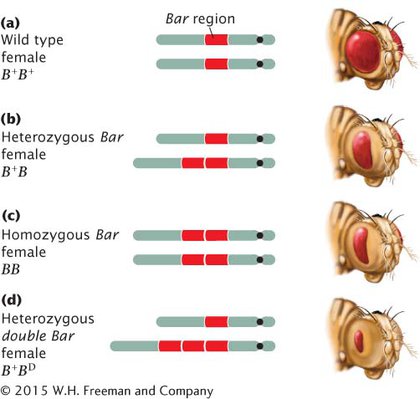
Duplications may have major effects on the phenotype. Among fruit flies, for example, a fly with the Bar mutation has a reduced number of facets in the eye, making the eye smaller and bar shaped instead of oval (Figure 6.6). The Bar mutation results from a small duplication on the X chromosome that is inherited as an incompletely dominant, X-
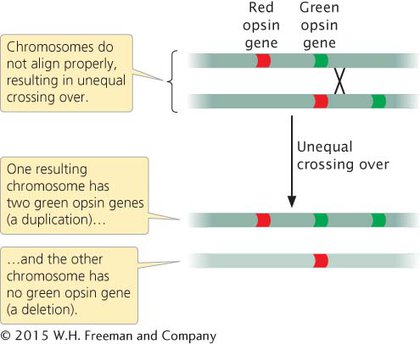
Duplications and deletions often arise from unequal crossing over, in which duplicated segments of chromosomes misalign during the process. Unequal crossing over is frequently the cause of red–green color blindness in humans. Perception of color is affected by red and green opsin genes, which are found on the X chromosome and are 98% identical in their DNA sequence. Most people with normal color vision have one red opsin gene and one green opsin gene (although some people have more than one copy of each). Because the red and green opsin genes are similar in sequence, they occasionally pair in prophase I, and unequal crossing over takes place (Figure 6.7; see also Figure 13.14). The unequal crossing over produces one chromosome with an extra opsin gene and one chromosome that is missing an opsin gene. When a male inherits the chromosome that is missing one of the opsin genes, red–green color blindness results.
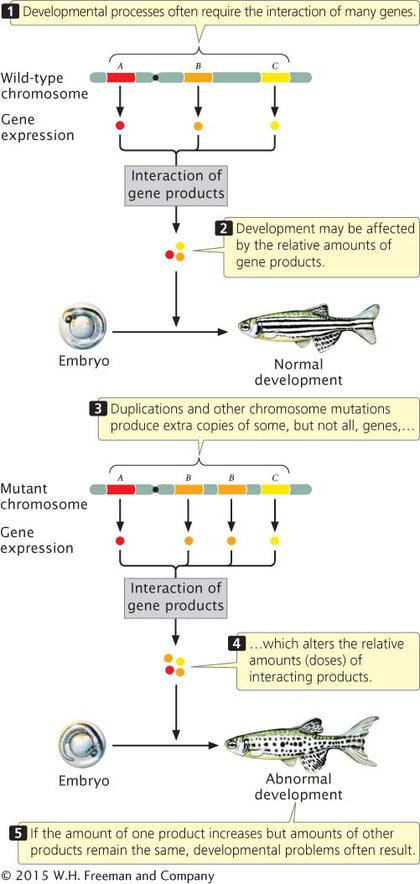
UNBALANCED GENE DOSAGE How does a chromosome duplication alter the phenotype? After all, gene sequences are not usually altered by duplications, and no genetic information is missing; the only change is the presence of additional copies of normal sequences. The answer to this question is not well understood, but the effects are most likely due to imbalances in the amounts of gene products (abnormal gene dosage). The amount of a particular protein synthesized by a cell is often directly related to the number of copies of its corresponding gene: an individual organism with three functional copies of a gene often produces 1.5 times as much of the protein encoded by that gene as an individual with two copies. Because developmental processes require the interaction of many proteins, they often depend critically on proper gene dosage. If the amount of one protein increases while the amounts of others remain constant, problems can result (Figure 6.8). Duplications can have severe consequences when the precise balance of a gene product is critical to cell function (Table 6.1).
| Type of rearrangement | Chromosome | Disorder | Symptoms |
|---|---|---|---|
| Duplication | 4, short arm | — | Small head, short neck, low hairline, reduced growth, and intellectual disability |
| Duplication | 4, long arm | — | Small head, sloping forehead, hand abnormalities |
| Duplication | 7, long arm | — | Delayed development, asymmetry of the head, fuzzy scalp, small nose, low- |
| Duplication | 9, short arm | — | Characteristic face, variable intellectual disability, high and broad forehead, hand abnormalities |
| Deletion | 5, short arm | Cri- |
Small head, distinctive cry, widely spaced eyes, round face, intellectual disability |
| Deletion | 4, short arm | Wolf–Hirschhorn syndrome | Small head with high forehead, wide nose, cleft lip and palate, severe intellectual disability |
| Deletion | 4, long arm | — | Small head, mild to moderate intellectual disability, cleft lip and palate, hand and foot abnormalities |
| Deletion | 7, long arm | Williams–Beuren syndrome | Facial features, heart defects, mental impairment |
| Deletion | 15, long arm | Prader–Willi syndrome | Feeding difficulty at early age but becoming obese after 1 year of age, mild to moderate intellectual disability |
| Deletion | 18, short arm | — | Round face, large low- |
| Deletion | 18, long arm | — | Distinctive mouth shape, small hands, small head, intellectual disability |
SEGMENTAL DUPLICATIONS The human genome contains numerous duplicated sequences called segmental duplications, which are defined as duplications greater than 1000 base pairs (1000 bp) in length. In most segmental duplications, the two copies are found on the same chromosome (an intrachromosomal duplication), but in others, the two copies are found on different chromosomes (an interchromosomal duplication). Many segmental duplications have been detected with the use of molecular techniques that examine the structure of DNA sequences on a chromosome (see Chapter 15). These techniques reveal that about 4% of the human genome consists of segmental duplications. In the human genome, the average size of segmental duplications is 15,000 bp.
IMPORTANCE OF DUPLICATIONS IN EVOLUTION Chromosome variations are potentially important in evolution, and they have clearly played a significant role in past evolution within a number of different groups of organisms. Chromosome duplications provide one possible way in which new genes evolve. In many cases, existing copies of a gene are not free to vary because they encode a product that is essential to development or function. However, after a chromosome undergoes duplication, extra copies of genes within the duplicated region are present. The original copy can provide the essential function, whereas an extra copy from the duplication is potentially free to undergo mutation and change. Over evolutionary time, the extra copy may acquire enough mutations to assume a new function that benefits the organism. For example, humans have a series of genes that encode different globin chains, which function as oxygen carriers. Some of these globin chains function during adult stages, and others function during embryonic and fetal development. The genes encoding all of these globins arose from an original ancestral gene that underwent a series of duplications.
CONCEPTS
A chromosome duplication is a mutation that doubles part of a chromosome. In individuals heterozygous for a chromosome duplication, the duplicated region of the chromosome loops out when homologous chromosomes pair in prophase I of meiosis. Duplications often have major effects on the phenotype, possibly by altering gene dosage. Segmental duplications are common within the human genome and have played an important role in the evolution of many eukaryotes.
 CONCEPT CHECK 1
CONCEPT CHECK 1
Chromosome duplications often result in abnormal phenotypes because
developmental processes depend on the relative amounts of proteins encoded by different genes.
extra copies of the genes within the duplicated region do not pair in meiosis.
the chromosome is more likely to break when it loops in meiosis.
extra DNA must be replicated, which slows down cell division.
a