Inversions
A third type of chromosome rearrangement is a chromosome inversion, in which a chromosome segment is inverted—
Inversion heterozygotes are common in many organisms, including a number of plants, some species of Drosophila, mosquitoes, and grasshoppers. Inversions may have played an important role in human evolution: G-
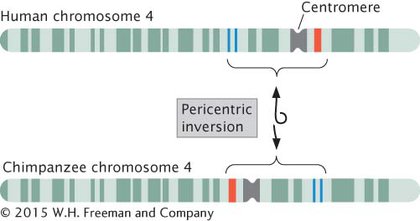
EFFECTS OF INVERSIONS Individual organisms with inversions have neither lost nor gained any genetic material; only the order of the chromosome segment has been altered. Nevertheless, these mutations often have pronounced phenotypic effects. An inversion may break a gene into two parts, and one part may move to a new location and destroy the function of the gene in that location. Even when the chromosome breaks lie between genes, phenotypic effects may arise from the inverted gene order. Many genes are regulated in a position-
INVERSIONS IN MEIOSIS When an individual is homozygous for a particular inversion, no special problems arise in meiosis, and the two homologous chromosomes can pair and separate normally. However, when an individual is heterozygous for an inversion, the gene order of the two homologs differs, and the homologous sequences can align and pair only if the two chromosomes form an inversion loop (Figure 6.12).
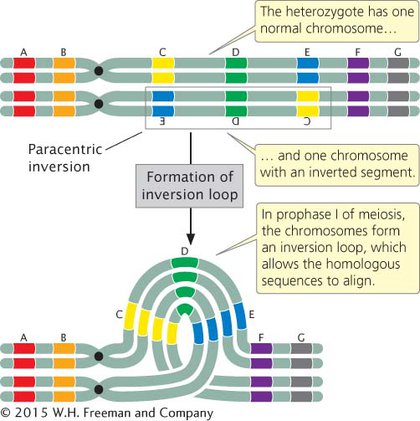
Individuals heterozygous for inversions also exhibit reduced recombination among genes located in the inverted region. The frequency of crossing over within the inversion is not actually diminished, but when crossing over does take place, the result is abnormal gametes that do not give rise to viable offspring, and thus no recombinant progeny are observed. Let’s see why this happens.
Figure 6.13 illustrates the results of crossing over within a paracentric inversion. The individual is heterozygous for an inversion (see Figure 6.13a), with one wild-
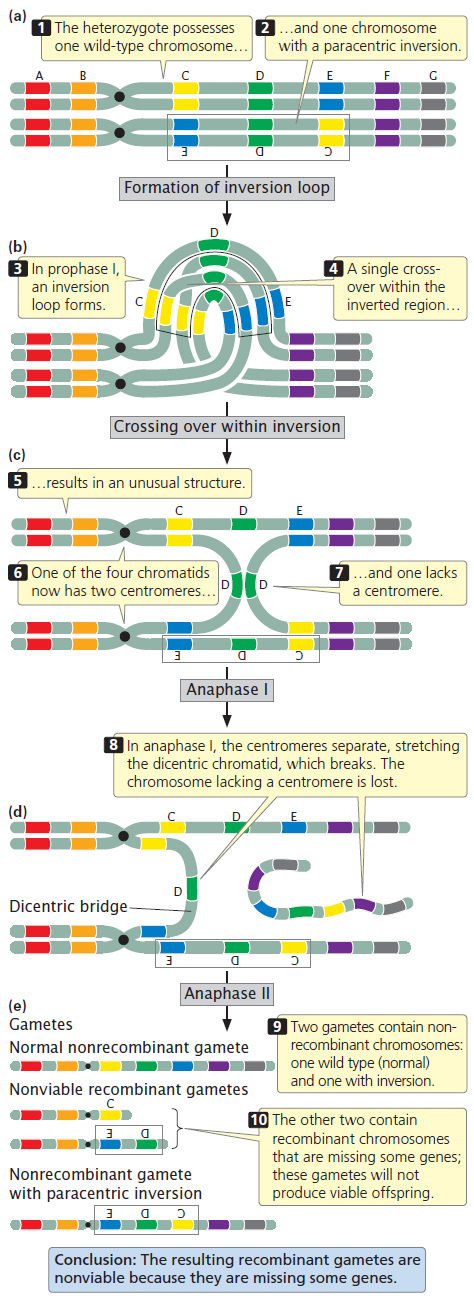
In anaphase I of meiosis, the centromeres are pulled toward opposite poles and the two homologous chromosomes separate. This action stretches the dicentric chromatid across the center of the nucleus, forming a structure called a dicentric bridge (see Figure 6.13d). Eventually, the dicentric bridge breaks as the two centromeres are pulled farther apart. Spindle microtubules do not attach to the acentric fragment, so this fragment does not segregate to a spindle pole and is usually lost when the nucleus re-
In the second division of meiosis, the chromatids separate and four gametes are produced (see Figure 6.13e). Two of the gametes contain the original, nonrecombinant chromosomes (AB•CDEFG and AB•EDCFG). The other two gametes contain recombinant chromosomes that are missing some genes; these gametes will not produce viable offspring. Thus, no recombinant progeny result when crossing over takes place within a paracentric inversion. The key is to recognize that crossing over still takes place, but when it does so, the resulting recombinant gametes are not viable, so no recombinant progeny are observed.
Recombination is also reduced within a pericentric inversion (Figure 6.14). No dicentric bridges or acentric fragments are produced, but the recombinant chromosomes have too many copies of some genes and no copies of others, so gametes that receive the recombinant chromosomes cannot produce viable progeny.
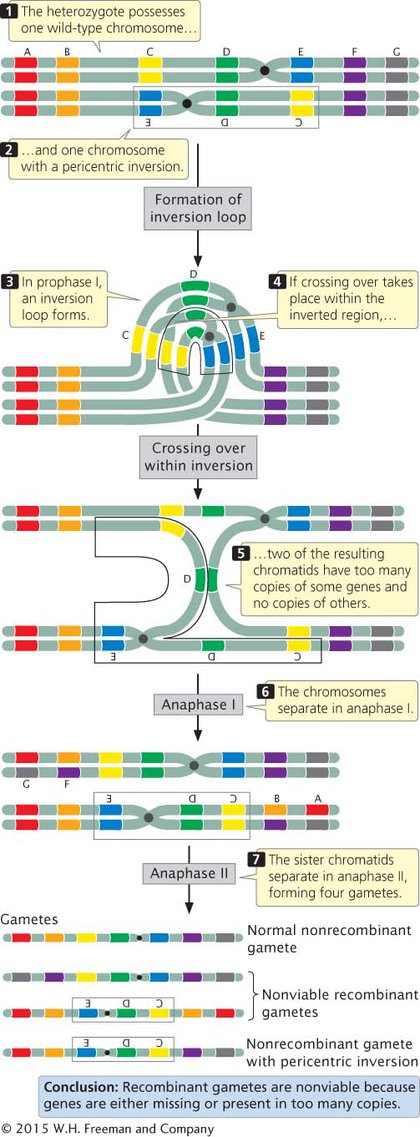
Figure 6.14 illustrates the results of single crossovers within inversions. Double crossovers in which both crossovers are on the same two strands (two- TRY PROBLEM 19
TRY PROBLEM 19
IMPORTANCE OF INVERSIONS IN EVOLUTION Inversions can also play important evolutionary roles by suppressing recombination among a set of genes. As we have seen, crossing over within an inversion in an individual heterozygous for a pericentric or paracentric inversion leads to unbalanced gametes and no recombinant progeny. This suppression of recombination allows particular combinations of alleles that function well together to remain intact, unshuffled by recombination.
CONCEPTS
In an inversion, a segment of a chromosome is turned 180 degrees. Inversions cause breaks in some genes and may move others to new locations. In individuals heterozygous for a chromosome inversion, the homologous chromosomes form an inversion loop in prophase I of meiosis. When crossing over takes place within the inverted region, nonviable gametes are usually produced, resulting in a depression in observed recombination frequencies.
 CONCEPT CHECK 3
CONCEPT CHECK 3
A dicentric chromosome is produced when crossing over takes place in an individual heterozygous for which type of chromosome rearrangement?
Duplication
Deletion
Paracentric inversion
Pericentric inversion
c