CONCEPT15.6 Recombination, Lateral Gene Transfer, and Gene Duplication Can Result in New Features
Several evolutionary processes can result in the acquisition of major new characteristics in populations. Each of these processes results in larger and more rapid evolutionary changes than do single point mutations.
Sexual recombination amplifies the number of possible genotypes
In asexually reproducing organisms, each new individual is genetically identical to its parent unless there has been a mutation. When organisms reproduce sexually, however, offspring differ from their parents because of crossing over and independent assortment of chromosomes during meiosis, as well as the combination of genetic material from two different gametes, as described in Concept 7.4. Sexual recombination generates an endless variety of genotype combinations that increase the evolutionary potential of populations—a long-term advantage of sex. Although some species may reproduce asexually most of the time, most asexual species have some means of achieving genetic recombination.
The evolution of meiosis and sexual recombination was a crucial event in the history of life. Exactly how these processes arose is puzzling, however, because in the short term, sex has at least three striking disadvantages:
- Recombination breaks up adaptive combinations of genes.
- Sex reduces the rate at which females pass genes on to their offspring.
- Dividing offspring into separate genders greatly reduces the overall reproductive rate.
319
To see why this last disadvantage exists, consider an asexual female that produces the same number of offspring as a sexual female. Assume that both females produce two offspring, but that half of the sexual female’s offspring are males. In the next (F1) generation, then, each of the two asexual F1 females will produce two more offspring—but there is only one sexual F1 female to produce offspring. Thus the effective reproductive rate of the asexual lineage is twice that of the sexual lineage. The evolutionary problem is to identify the advantages of sex that can overcome such short-term disadvantages.
A number of hypotheses have been proposed to explain the existence of sex, none of which are mutually exclusive. One is that sexual recombination facilitates repair of damaged DNA, because breaks and other errors in DNA on one chromosome can be repaired by copying the intact sequence from the homologous chromosome.
Another advantage of sexual reproduction is that it permits the elimination of deleterious mutations through recombination followed by selection. As Concept 9.2 described, DNA replication is not perfect, and many replication errors result in lower fitness. Meiotic recombination distributes these deleterious mutations unequally among gametes. Sexual reproduction then produces some individuals with more deleterious mutations and some with fewer. The individuals with fewer deleterious mutations are more likely to survive. Therefore sexual reproduction allows natural selection to eliminate particular deleterious mutations from the population over time.
In asexual reproduction, deleterious mutations can be eliminated only by the death of the lineage or by a rare back mutation (that is, when a subsequent mutation returns a mutated sequence to its original DNA sequence). Hermann J. Muller noted that deleterious mutations in a non-recombining genome accumulate—“ratchet up”—at each replication. Mutations occur and are passed on each time a genome replicates, and these mutations accumulate with each subsequent generation. This accumulation of deleterious mutations in lineages that lack genetic recombination is known as Muller’s ratchet.
Another explanation for the existence of sex is that the great variety of genetic combinations created in each generation can itself be advantageous. For example, genetic variation can be a defense against pathogens and parasites. Most pathogens and parasites have much shorter life cycles than their hosts and can rapidly evolve counter-adaptations to host defenses. Sexual recombination might give the host’s defenses a chance to keep up.
Sexual recombination does not directly influence the frequencies of alleles. Rather, it generates new combinations of alleles on which natural selection can act. It expands variation in quantitative characters by creating new genotypes. That is why artificial selection for bristle number in Drosophila (see Figure 15.7) resulted in flies with either more or fewer bristles than were present in the flies in the initial population.
Lateral gene transfer can result in the gain of new functions
The tree of life is usually visualized as a branching diagram, with each lineage diverging into two (or more) lineages over time, from one common ancestor to the millions of species that are alive today. Ancestral lineages divide into descendant lineages, and it is those speciation events that the tree of life captures. However, there are also processes that result in lateral gene transfer—the horizontal movement of individual genes, organelles, or fragments of genomes from one lineage to another. Some species may pick up fragments of DNA directly from the environment. A virus may pick up some genes from one host and transfer them to a new host when the virus becomes integrated into the new host’s genome. Hybridization between species also results in the lateral transfer of large numbers of genes.
Lateral gene transfer can be highly advantageous to the species that incorporates novel genes from a distant relative. Genes that confer antibiotic resistance, for example, are commonly transferred among different species of bacteria. Lateral gene transfer is another way, in addition to mutation and recombination, that species can increase their genetic variation.
The degree to which lateral gene transfer events occur in various parts of the tree of life is a matter of considerable current investigation and debate. Lateral gene transfer appears to be relatively uncommon among most eukaryote lineages, although the two major endosymbioses that gave rise to mitochondria and chloroplasts involved lateral transfers of entire bacterial genomes to the eukaryote lineage. Some groups of eukaryotes, most notably some plants, are subject to relatively high levels of hybridization among closely related species. Hybridization leads to the exchange of many genes among recently separated lineages of plants. The greatest degree of lateral transfer, however, occurs among bacteria. Many genes have been transferred repeatedly among bacteria, to the point that relationships and boundaries among species of bacteria are sometimes hard to decipher.
Many new functions arise following gene duplication
Gene duplication is yet another way that genomes can acquire new functions. When a gene is duplicated, one copy of that gene is potentially freed from having to perform its original function. The identical copies of a duplicated gene can have any one of four different fates:
- Both copies of the gene may retain their original function (which can result in a change in the amount of gene product that is produced by the organism).
- Both copies of the gene may retain the ability to produce the original gene product, but the expression of the genes may diverge in different tissues or at different times in development.
- One copy of the gene may be incapacitated by the accumulation of deleterious mutations and become a functionless pseudogene.
- One copy of the gene may retain its original function while the second copy changes and evolves a new function.
How often do gene duplications arise, and which of these four outcomes is most likely? Investigators have found that rates of gene duplication are fast enough for a yeast or Drosophila population to acquire several hundred duplicate genes over the course of a million years. They have also found that most of the duplicated genes that are still present in these organisms are very young. Many duplicated genes are lost from a genome within 10 million years—an eyeblink on an evolutionary time scale.
320
Many gene duplications affect only one or a few genes at a time, but in some cases entire genomes may be duplicated. When all the genes are duplicated, there are massive opportunities for new functions to evolve. That is exactly what seems to have happened during the course of vertebrate evolution. The genomes of the jawed vertebrates have four diploid sets of many major genes, which leads biologists to conclude that two genome-wide duplication events occurred in the ancestor of these species. These duplications allowed considerable specialization of individual vertebrate genes, many of which are now highly tissue-specific in their expression.
LINK
See Concept 14.4 for a discussion of the role of duplicated Hox genes in vertebrate evolution
Several successive rounds of duplication and sequence evolution may result in a gene family, a group of homologous genes with related functions, often arrayed in tandem along a chromosome. An example of a group of genes related by gene duplication is the globin gene family (FIGURE 15.23). Comparisons of the amino acid sequences among globins strongly suggest that this family of proteins arose via gene duplications.
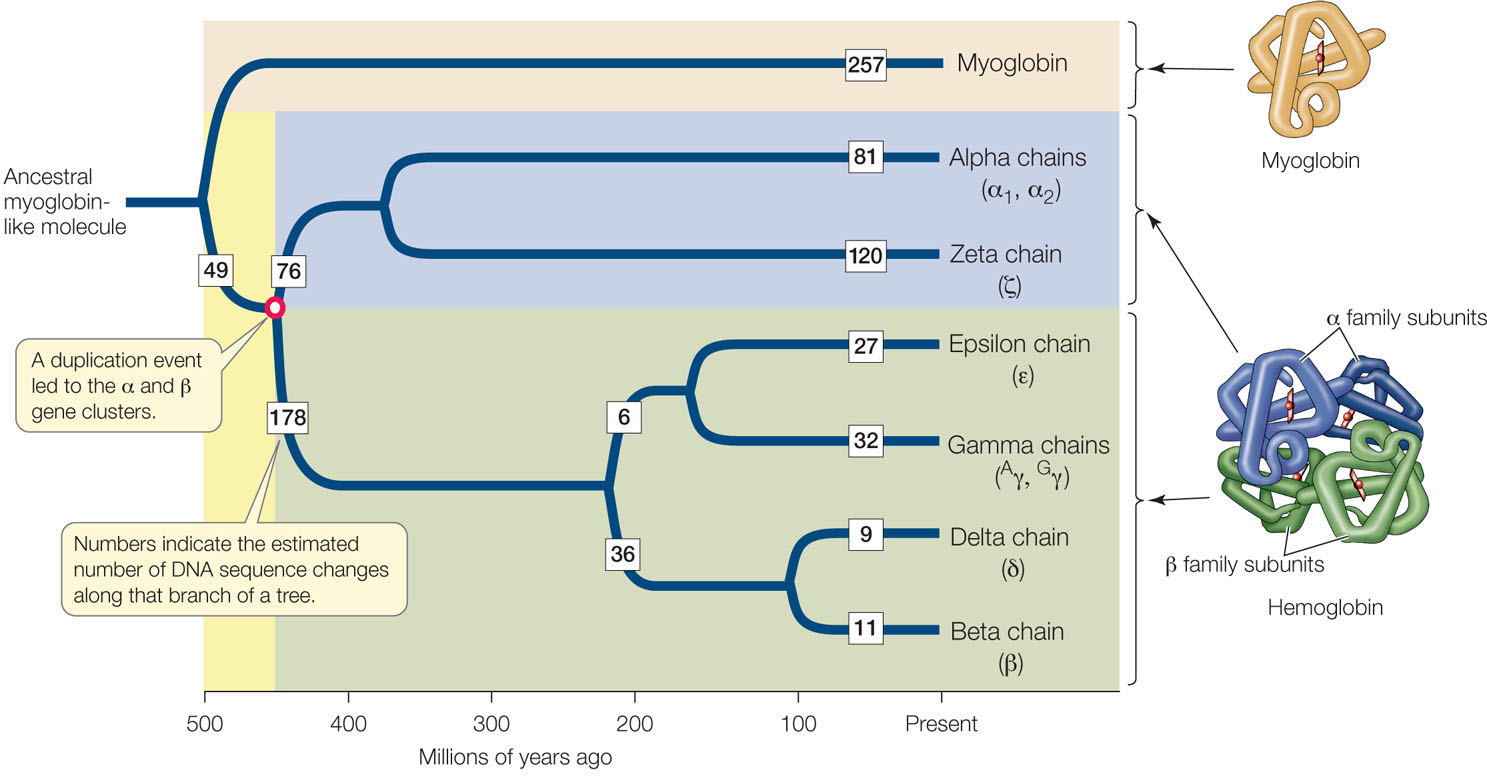
Hemoglobin is a tetramer (four-subunit molecule) consisting of two α-globin and two β-globin polypeptide chains. It carries oxygen in the blood. Myoglobin, a monomer, is the primary O2 storage protein in muscle. Myoglobin’s affinity for O2 is much higher than that of hemoglobin, but hemoglobin has evolved to be more diversified in its role. Hemoglobin binds O2 in the lungs or gills, where the O2 concentration is relatively high, transports it to deep body tissues, where the O2 concentration is low, and releases it in those tissues. With its more complex tetrameric structure, hemoglobin is able to carry four molecules of O2, as well as hydrogen ions and carbon dioxide, in the blood. Hemoglobin and myoglobin are estimated to have arisen through gene duplication about 500 million years ago.
CHECKpointCONCEPT15.6
- What are some of the potential advantages of lateral gene transfer to the organisms that gain new genes by this process?
- Why is gene duplication considered important for long-term evolutionary change?
- Why is sexual reproduction so prevalent in nature, despite its having at least three short-term evolutionary disadvantages?
The development of evolutionary theory has helped reveal how biological molecules function, how genetic diversity is created and maintained, and how organisms develop new features. Next we will see how biologists put this theory into practice.
321