CONCEPT16.4 Phylogeny Is the Basis of Biological Classification
The biological classification system in widespread use today is derived from that developed by the Swedish biologist Carolus Linnaeus in the mid-1700s. Linnaeus developed a system of binomial nomenclature. Linnaeus gave each species two names, one identifying the species itself and the other the group of closely related species, or genus (plural, genera), to which it belongs. Optionally, the name of the taxonomist who first proposed the species name may be added at the end. Thus Homo sapiens Linnaeus is the name of the modern human species. Homo is the genus, sapiens identifies the particular species in the genus Homo, and Linnaeus is the person who proposed the name Homo sapiens.
You can think of Homo as equivalent to your surname and sapiens as equivalent to your first name. The first letter of the genus name is capitalized, and the specific name is lowercase. Both of these formal designations are italicized. Rather than repeating the name of a genus when it is used several times in the same discussion, biologists often spell it out only once and abbreviate it with the initial letter thereafter (e.g., D. melanogaster rather than Drosophila melanogaster).
As we noted earlier, any group of organisms that is treated as a unit in a biological classification system, such as all species in the genus Drosophila, or all insects, or all arthropods, is called a taxon. In the Linnaean system, species and genera are further grouped into a hierarchical system of higher taxonomic categories. The taxon above the genus in the Linnaean system is the family. The names of animal families end in the suffix “-idae.” Thus Formicidae is the family that contains all ant species, and the family Hominidae contains humans and our recent fossil relatives, as well as our closest living relatives, the chimpanzees, gorillas, and orangutans. Family names are based on the name of a member genus; Formicidae is based on the genus Formica, and Hominidae is based on Homo. The same rules are used in classifying plants, except that the suffix “-aceae” is used for plant family names instead of “-idae.” Thus Rosaceae is the family that includes the genus Rosa (roses) and its relatives.
339
In the Linnaean system, families are grouped into orders, orders into classes, and classes into phyla (singular phylum), and phyla into kingdoms. However, the ranking of taxa within Linnaean classification is subjective. Whether a particular taxon is considered, say, an order or a class is informative only with respect to the relative ranking of other related taxa. Although families are always grouped within orders, orders within classes, and so forth, there is nothing that makes a “family” in one group equivalent (in number of genera or in evolutionary age, for instance) to a “family” in another group.
Today the Linnaean terms above the genus level are used largely for convenience. Linnaeus recognized the overarching hierarchy of life, but he developed his system before evolutionary thought had become widespread. Biologists today recognize the tree of life as the basis for biological classification and often name taxa without placing them into the various Linnaean ranks. But regardless of whether they rank organisms into Linnaean categories or use unranked taxon names, modern biologists use evolutionary relationships as the basis for distinguishing, naming, and classifying biological groups.
Evolutionary history is the basis for modern biological classification
Today’s biological classifications express the evolutionary relationships of organisms. Taxa are expected to be monophyletic, meaning that the taxon contains an ancestor and all descendants of that ancestor, and no other organisms. In other words, a monophyletic taxon is a historical group of related species, or a complete branch on the tree of life. As noted earlier, this is also the definition of a clade. A true monophyletic group can be removed from a phylogenetic tree by a single “cut” in the tree, as shown in FIGURE 16.12.
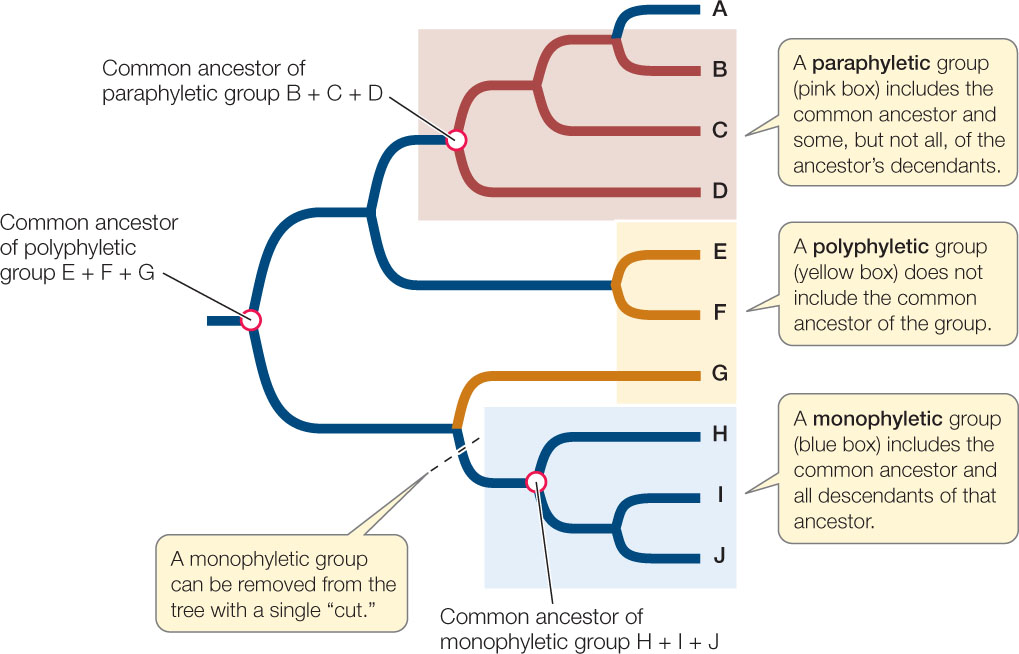
Note that there are many monophyletic groups on any phylogenetic tree, and that these groups are successively smaller subsets of larger monophyletic groups. This hierarchy of biological taxa, with all of life as the most inclusive taxon and many smaller taxa within larger taxa, down to the individual species, is the modern basis for biological classification.
Although biologists seek to describe and name only monophyletic taxa, the detailed phylogenetic information needed to do so is not always available. A group that does not include its common ancestor is polyphyletic. A group that does not include all the descendants of a common ancestor is referred to as paraphyletic (see Figure 16.12). Virtually all taxonomists now agree that polyphyletic and paraphyletic groups are inappropriate as taxonomic units because they do not correctly reflect evolutionary history. Some classifications still contain such groups because some organisms have not been evaluated phylogenetically. As mistakes in prior classifications are detected, taxonomic names are revised and polyphyletic and paraphyletic groups are eliminated from the classifications.
Several codes of biological nomenclature govern the use of scientific names
Several sets of explicit rules govern the use of scientific names. Biologists around the world follow these rules voluntarily to facilitate communication and dialogue. There may be dozens of common names for an organism in many different languages, and the same common name may refer to more than one species (FIGURE 16.13). The rules of biological nomenclature are designed so that there is only one correct scientific name for any single recognized taxon, and (ideally) a given scientific name applies only to a single taxon (that is, each scientific name is unique). Sometimes the same species is named more than once (when more than one taxonomist has taken up the task). In these cases, the rules specify that the valid name is the first name that was proposed. If the same name is inadvertently given to two different species, then the species that was named second must be given a new name.

340
Phylogeny is the basis of biological classification
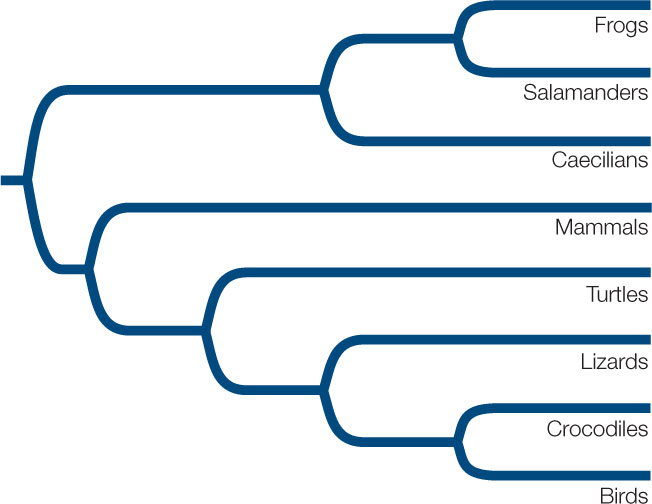
Consider the above phylogeny and three possible classifications of the living taxa.
- Which of these classifications contains a paraphyletic group?
- Which of these classifications contains a polyphyletic group?
- Which of these classifications is consistent with the goal of including only monophyletic groups in a biological classification?
- Starting with the classification you named in Question 3, how many additional group names would you need to include all the clades shown in this phylogenetic tree?
341
Because of the historical separation of the fields of zoology, botany (which originally included mycology, the study of fungi), and microbiology, different sets of taxonomic rules were developed for each of these groups. Yet another set of rules emerged later for classifying viruses. This separation of fields resulted in duplicated taxon names in groups governed by the different sets of rules. Drosophila, for example, is both a genus of fruit flies and a genus of fungi, and some species in both groups have identical names. Until recently these duplicated names caused little confusion, since traditionally biologists who studied fruit flies were unlikely to read the literature on fungi (and vice versa). Today, given the prevalence of large, universal biological databases (such as GenBank, which includes DNA sequences from across all life), it is increasingly important that each taxon have a unique and unambiguous name. Biologists are working on a universal code of nomenclature that can be applied to all organisms, so that every species will have a unique identifying name or registration number. This will assist efforts to build an online Encyclopedia of Life that links all the information for all the world’s species.
CHECKpointCONCEPT16.4
- What is the difference between monophyletic, paraphyletic, and polyphyletic groups?
- Why do biologists prefer monophyletic groups in formal classifications?
- What advantages or disadvantages do you see to having separate sets of taxonomic rules for animals, plants, bacteria, and viruses?
Having described some of the mechanisms by which evolution occurs and how phylogenies can be used to study evolutionary relationships, we are now ready to consider the subject of speciation. Speciation is the process that leads to the splitting events (nodes) on the tree of life and eventually results in the millions of distinctive species that constitute Earth’s biodiversity.
How are phylogenetic methods used to resurrect protein sequences from extinct organisms?
ANSWER Most genes and proteins of organisms that lived millions of years ago have decomposed in the fossil remains of these species. Nonetheless, sequences of many ancient genes and proteins can be reconstructed. Just as we can reconstruct the morphological features of a clade’s ancestors, we can also reconstruct the ancestors’ DNA and protein sequences—if we have enough information about the genomes of their descendants (Concept 16.3).
Biologists have now reconstructed gene sequences from many species that have been extinct for millions of years. Using this information, a laboratory can reconstruct real proteins that correspond to the proteins that were present in long-extinct species. This is how Mikhail Matz and his colleagues were able to resurrect fluorescent proteins from the extinct ancestors of modern corals and then visualize the colors produced by these proteins in the laboratory and recreate the probable evolutionary path of the different pigments (FIGURE 16.14).
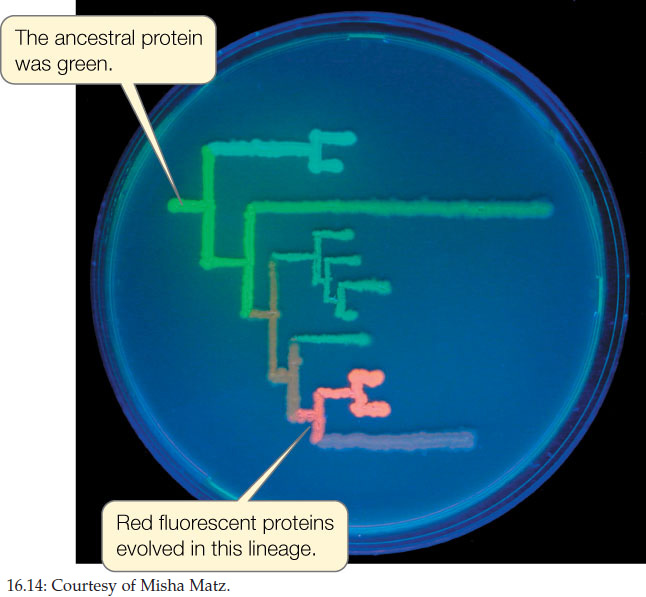
Biologists have even used phylogenetic analysis to reconstruct some protein sequences that were present in the common ancestor of life (Concept 16.1). These hypothetical protein sequences can then be resurrected into actual proteins in the laboratory. When biologists measured the temperature optima for these resurrected proteins, they found that the proteins functioned best in the range of 55°C–65°C. This analysis is consistent with hypotheses that life evolved in a high-temperature environment.
To reconstruct protein sequences from species that have been extinct for millions or even billions of years, biologists use detailed mathematical models that take into account much of what we have learned of molecular evolution (Concept 16.2). These models incorporate information on rates of replacement among different amino acid residues in proteins, information on different substitution rates among nucleotides, and changes in the rate of molecular evolution among the major lineages of life.
342