DNA AND DESCENT
While all living organisms share DNA and the genetic code, no two species share the exact same sequence of DNA nucleotides. That’s because (as described in Chapter 10) errors in DNA replication and other mutations are continually introducing variation into DNA sequences (and the proteins they encode). Over time, neutral and advantageous mutations will tend to be preserved, while harmful mutations will tend to be selected against and eliminated. In addition, much of our DNA consists of long stretches of noncoding sequences with no known function. Because mutations in these regions have no effect on an organism, they accumulate over time. As mutations are passed on to descendants, the number of sequence differences between the ancestor and its descendants grows—slowly in the case of sequences coding for critical proteins whose structures are well adapted to their functions, and more rapidly in the case of noncoding DNA not involved with making proteins. Closely related species will therefore have more similar DNA sequences than species that are more distantly related.
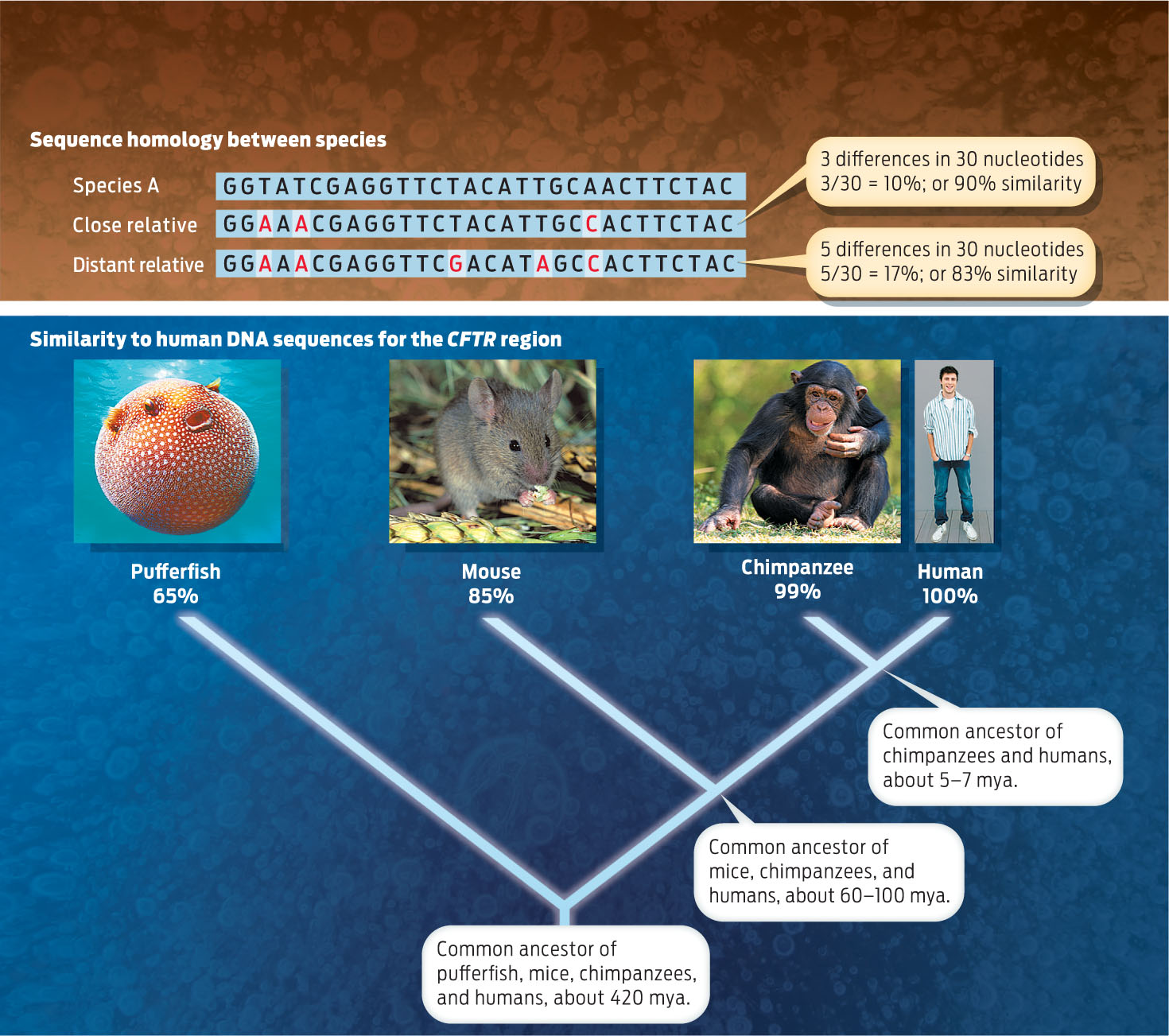
For example, when scientists looked at one specific region of DNA—the cystic fibrosis transmembrane regulator (or CFTR) region—they discovered that human DNA in this region is 99% identical to chimpanzee DNA. The fact that the DNA of the two species is nearly identical reflects the fact that humans and chimps share a common ancestor that lived relatively recently—just 5–7 million years ago. By contrast, human DNA is 85% identical to the DNA of a mouse at this same region, which makes sense given that humans and mice share a common ancestor that lived between 60 and 100 million years ago. Less sequence identity would be seen between a human and a toad, whose common ancestor—a lobed-finned fish—lived roughly 375 million years ago. The more distantly related two species are, the more sequence differences in DNA sequences you will see. In essence, DNA serves as a kind of molecular clock: each additional sequence difference is like a tick of the clock, showing the amount of time that has elapsed since the two species shared a common ancestor (INFOGRAPHIC 16.7).
Related organisms share DNA sequences inherited from a common ancestor. Over time, the sequence in each species acquires independent mutations. The more time that has passed, the greater the number of sequence differences that will be present. Thus, the percentage of nucleotides that differ between two species gives an indication of the evolutionary distance between them.
When combined with evidence from the fossil record, anatomy, and development, molecular data become a powerful tool for understanding evolution. As we’ll see in Chapter 17, DNA evidence is often a more reliable clue to common ancestry than physical appearance, and can serve as a check on conclusions derived from the fossil record or anatomy. As well, DNA is deepening our knowledge of how limbs are constructed at the molecular level. Scientists working in Shubin’s lab have shown that the same genes orchestrate limb development in animals as diverse as sharks, chickens, and humans. Learning how these genes work and how changes in their DNA sequences can produce large-scale changes in body plan or limb structure is a hot area of biology right now, informally known as “evo-devo” (short for “evolutionary developmental biology”).