12.3 Proteins Carry Out Most Membrane Processes
✓ 3 Describe the roles of proteins in making membranes selectively permeable.
We now turn to membrane proteins, which are responsible for most of the dynamic processes carried out by membranes. Membrane lipids form a permeability barrier and thereby establish compartments, whereas specific proteins mediate nearly all other membrane functions. In particular, proteins transport chemicals and information across a membrane. Membranes that differ in function differ in their protein content. Myelin, a membrane that serves as an electrical insulator around certain nerve fibers, has a low content of protein (18%). Membranes composed almost entirely of lipids are suitable for insulation because the hydrophobic components do not conduct currents well. In contrast, the plasma membranes, or exterior membranes, of most other cells must conduct the traffic of molecules into and out of the cells and so contain many pumps, channels, receptors, and enzymes. The protein content of these plasma membranes is typically 50%. Energy-
Proteins Associate with the Lipid Bilayer in a Variety of Ways
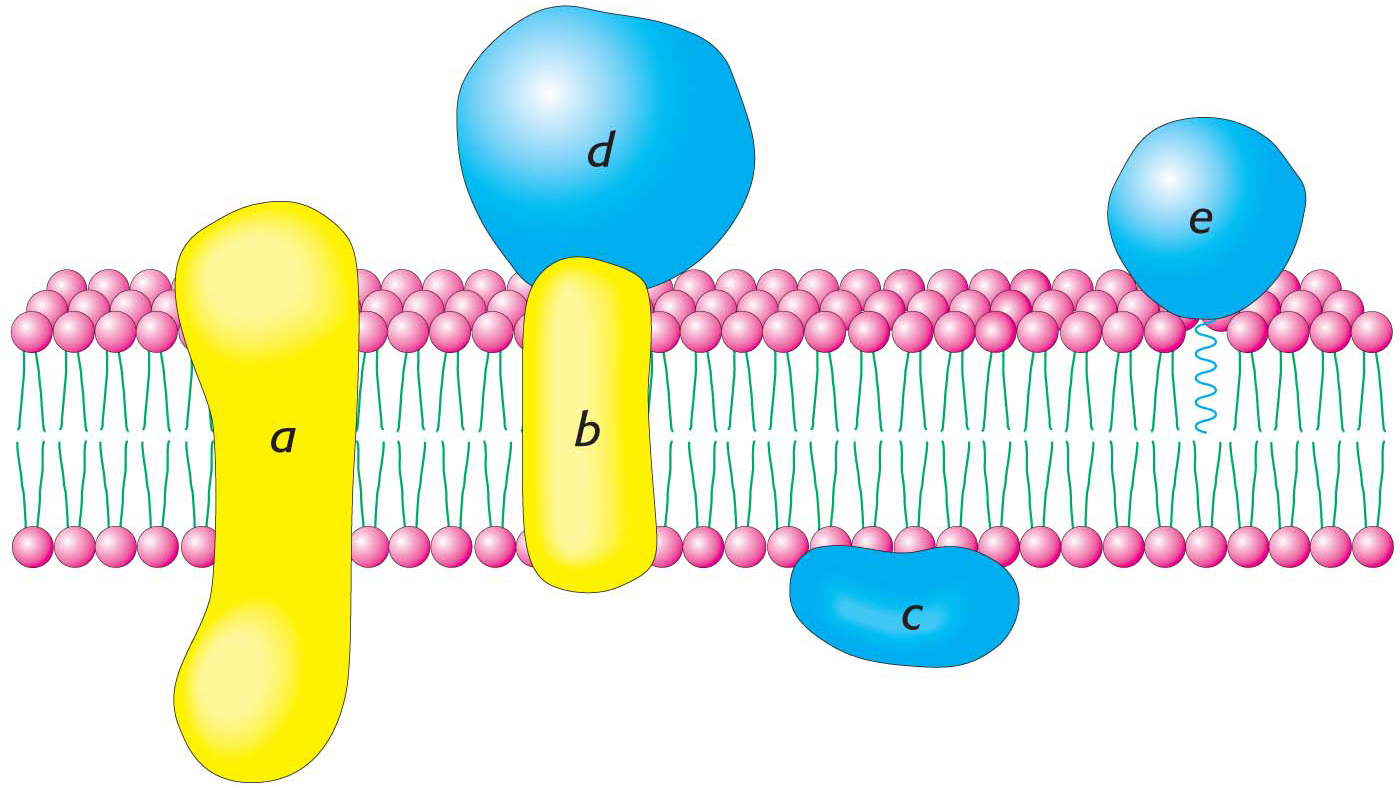
Membrane proteins can be classified as being either peripheral or integral on the basis of their interaction with the hydrophobic interior of the membrane (Figure 12.8). Integral membrane proteins are embedded in the hydrocarbon chains of membrane lipids, and they can be released only when the membrane is physically disrupted. In fact, most integral membrane proteins span the lipid bilayer. In contrast, peripheral membrane proteins are bound to the head groups of lipids or the exposed portions of integral membrane proteins by electrostatic and hydrogen-
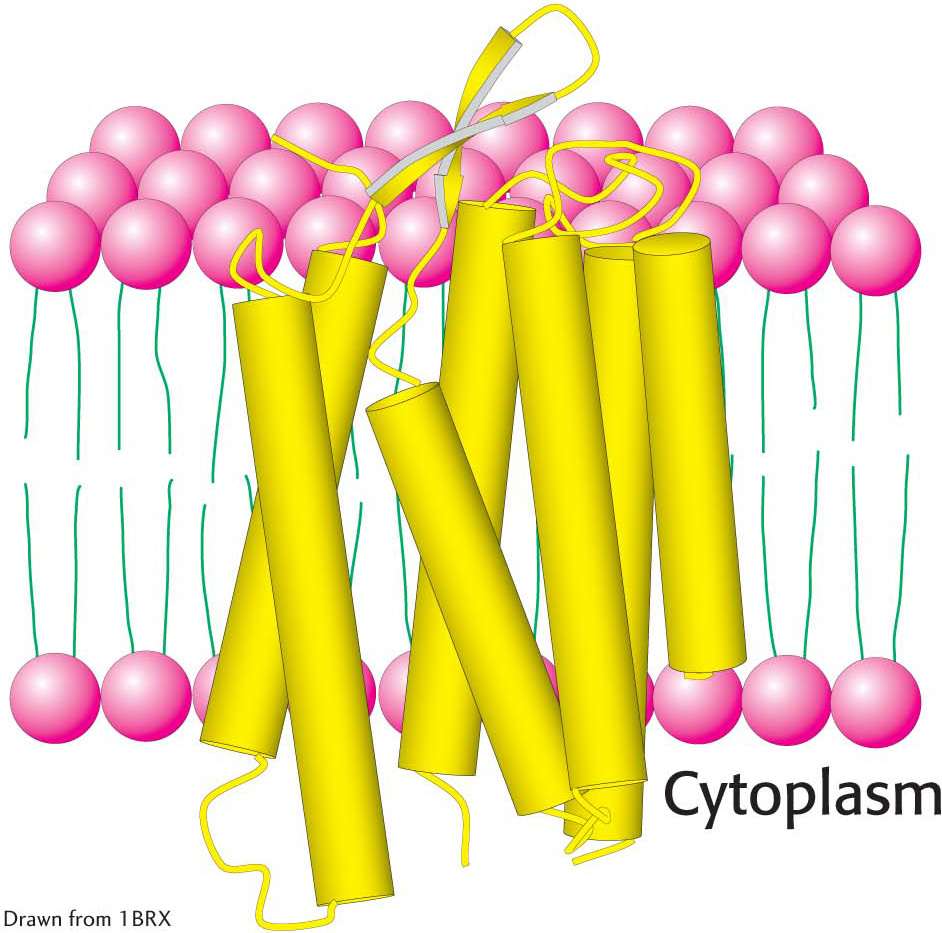
Proteins can span the membrane with α helices. For instance, consider bacteriorhodopsin, which uses light energy to transport protons from inside the bacterial cell to outside, generating a proton gradient used to form ATP (Figure 12.9). Bacteriorhodopsin is built almost entirely of α helices; seven closely packed α helices, arranged almost perpendicularly to the plane of the cell membrane, span its width. Just as the nonpolar hydrocarbon moieties of phospholipids associate with one another, the nonpolar α helices of bacteriorhodopsin associate with the hydrocarbon core of the lipid bilayer. Membrane-
The membrane-
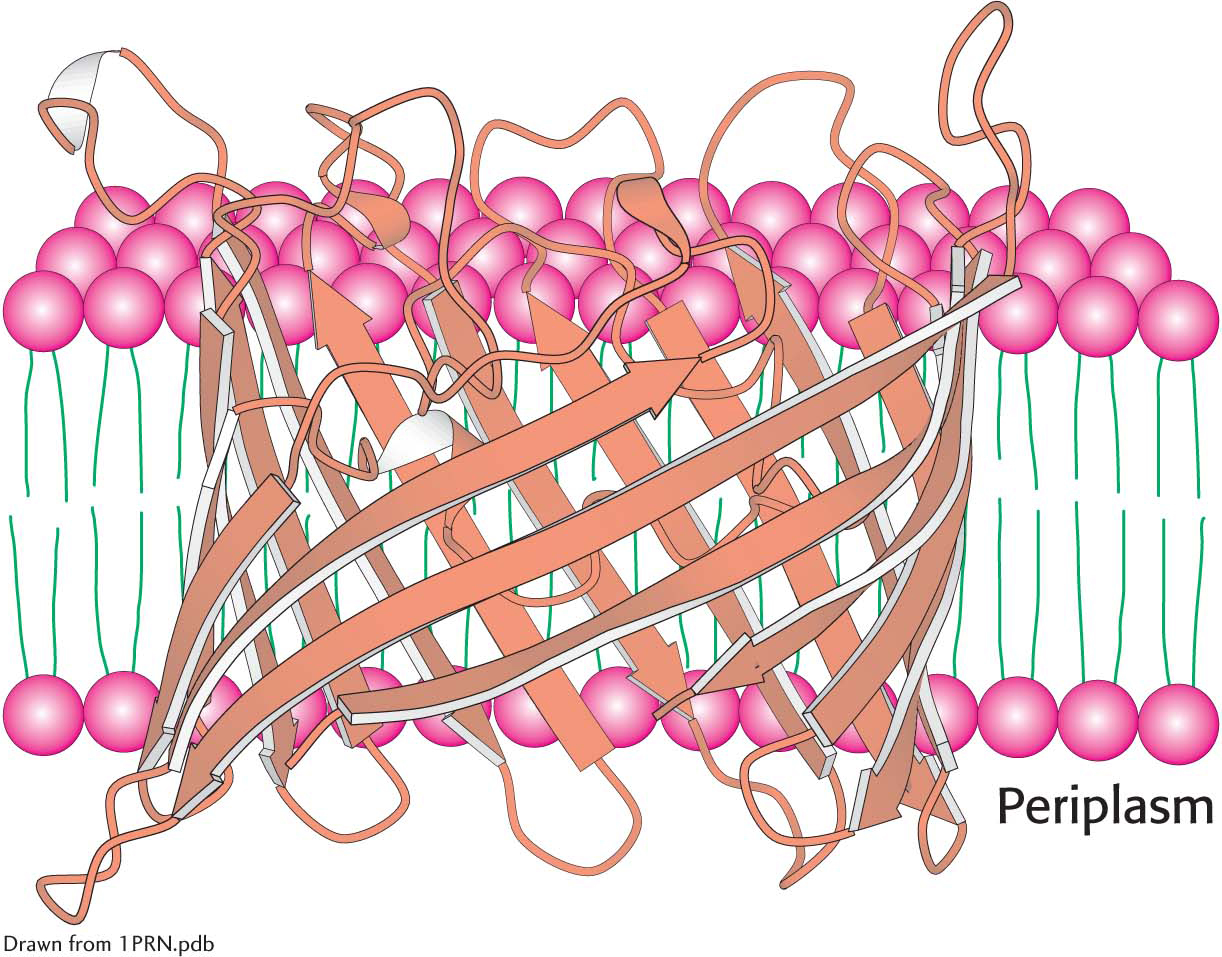
Another means by which a protein associates with a membrane is the embedding of just part of the protein into the membrane. The structure of the endoplasmic reticulum membrane-
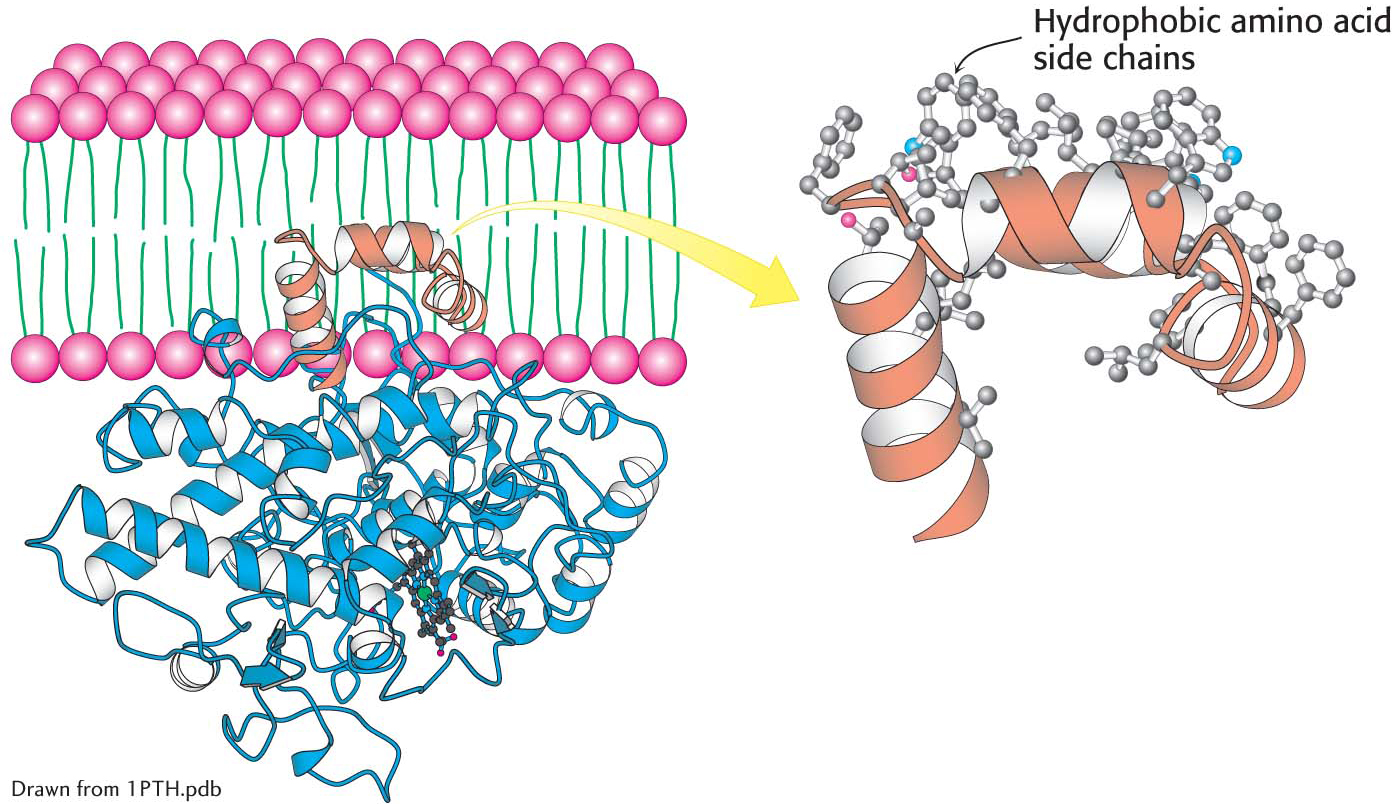
 CLINICAL INSIGHT
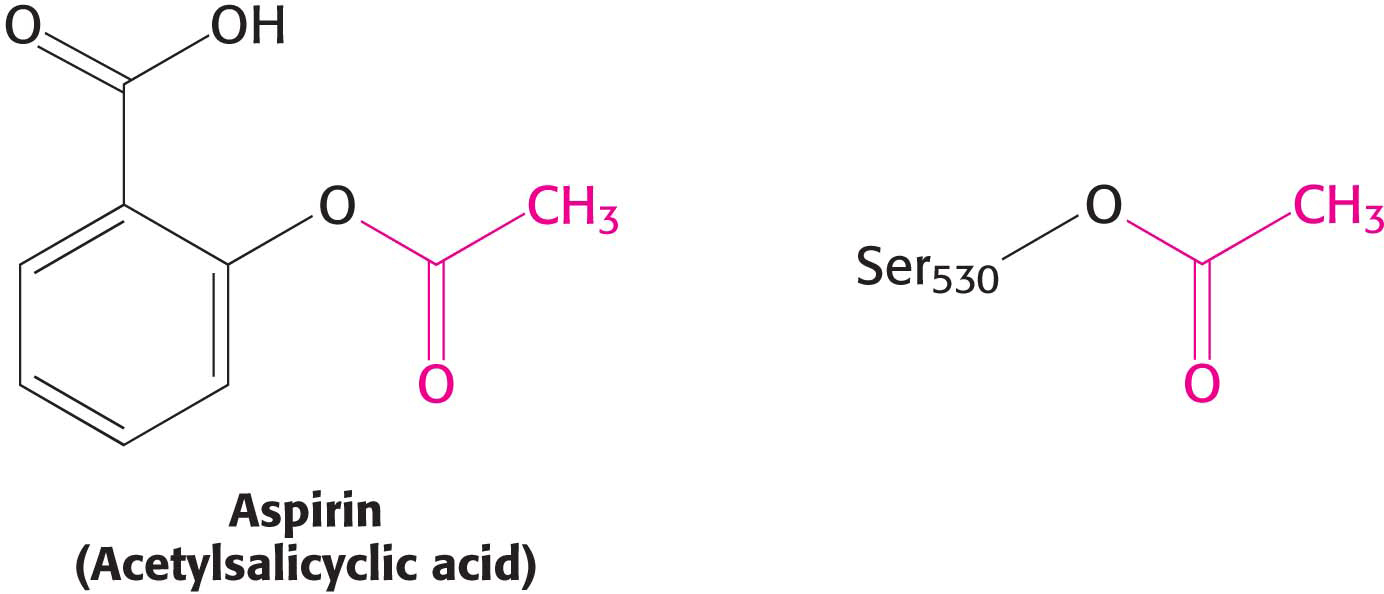
CLINICAL INSIGHTThe Association of Prostaglandin H2 Synthase-l with the Membrane Accounts for the Action of Aspirin
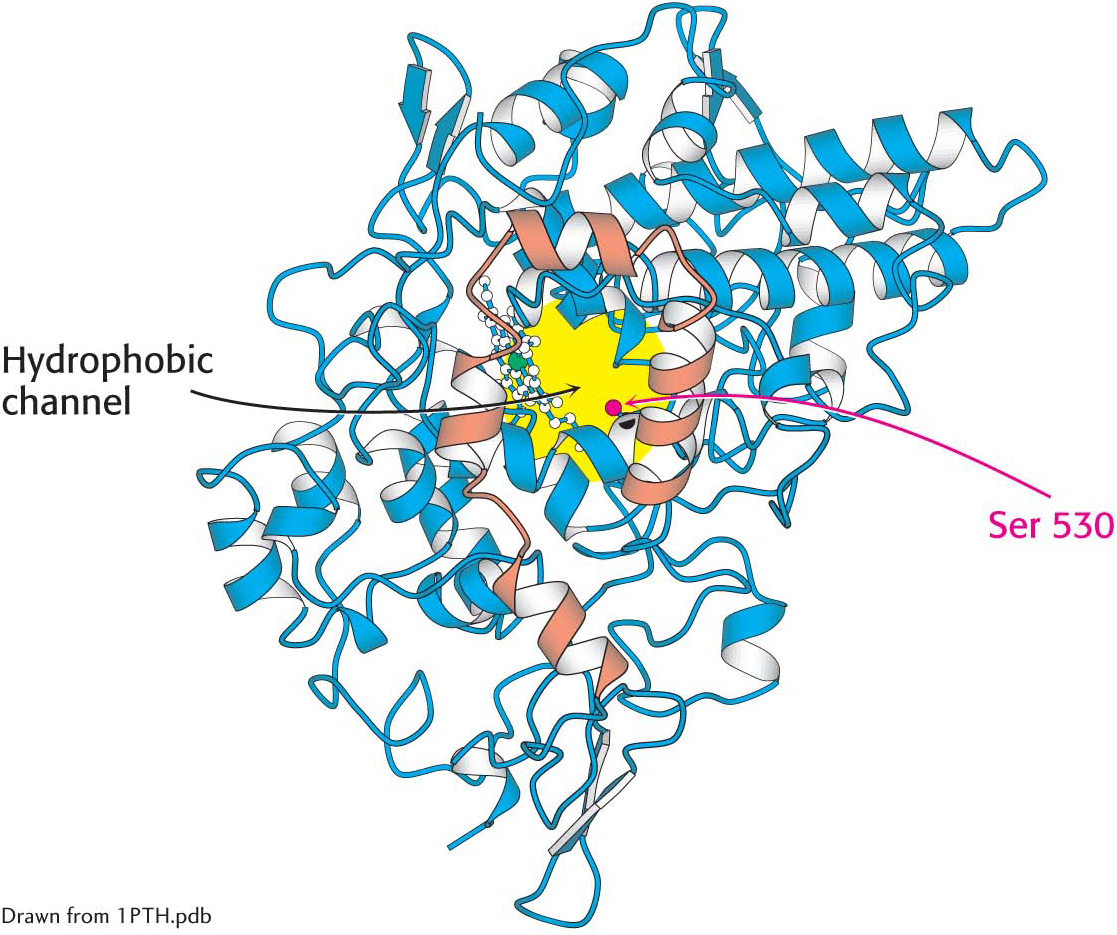
The localization of prostaglandin H2 synthase-