
34.1 DNA Is Replicated by Polymerases
✓ 3 Identify the enzymes that take part in the process of DNA replication.
The full replication machinery in a cell comprises more than 20 proteins engaged in intricate and coordinated interplay. The key enzymes are called DNA polymerases, which promote the formation of the phosphodiester linkages joining units of the DNA backbone. E. coli has five DNA polymerases, designated by roman numerals, that participate in DNA replication and repair (Table 34.1). We will focus our attention on two of the better understood polymerases: DNA polymerase I and DNA polymerase III.

DNA Polymerase Catalyzes Phosphodiester-Linkage Formation
DNA polymerases catalyze the step-

where dNTP stands for any deoxyribonucleotide and PPi is a pyrophosphate ion. DNA synthesis has the following characteristics:
DID YOU KNOW?
A template is a sequence of nucleic acids that determines the sequence of a complementary nucleic acid.
DID YOU KNOW?
A primer is the initial segment of a polymer that is to be extended on which elongation depends.
The reaction requires all four activated precursors—
that is, the deoxynucleoside 5′-triphosphates dATP, dGTP, dCTP, and TTP—as well as the Mg2+ ion. The new DNA strand is assembled directly on a preexisting DNA template. DNA polymerases catalyze the formation of a phosphodiester linkage efficiently only if the base on the incoming nucleoside triphosphate is complementary to the base on the template strand. Thus, DNA polymerase is a template-
directed enzyme that synthesizes a product with a base sequence complementary to that of the template.Page 629 Figure 34.1 A polymerization reaction catalyzed by DNA polymerases.
Figure 34.1 A polymerization reaction catalyzed by DNA polymerases.DNA polymerases require a primer to begin synthesis. A primer strand having a free 3′-OH group must be already bound to the template strand. The strand-
elongation reaction catalyzed by DNA polymerases is a nucleophilic attack by the 3′-OH end of the growing strand on the innermost phosphorus atom of deoxynucleoside triphosphate (Figure 34.2). A phosphodiester bridge is formed and pyrophosphate is released. The subsequent hydrolysis of pyrophosphate to yield two ions of orthophosphate (Pi) by pyrophosphatase drives the polymerization forward. Elongation of the DNA strand proceeds in the 5′-to- 3 ′ direction. Figure 34.2 Strand-
Figure 34.2 Strand-elongation reaction . DNA polymerases catalyze the formation of a phosphodiester bridge. Elongation of the DNA strand proceeds in the 5′-to-3′ direction. Many DNA polymerases are able to correct mistakes in DNA by removing mismatched nucleotides. These polymerases have a distinct nuclease activity that allows them to excise incorrect bases by a separate reaction. For instance, DNA polymerase I has three distinct active sites: the polymerase site, a 3′ → 5′ exonuclease site, and a 5′ → 3′ exonuclease site. The 3′ → 5′ nuclease activity contributes to the remarkably high fidelity of DNA replication, which has an error rate of less than 10−8 per base pair. We will consider the function of the 5′ → 3′ nuclease activity shortly.
The three-
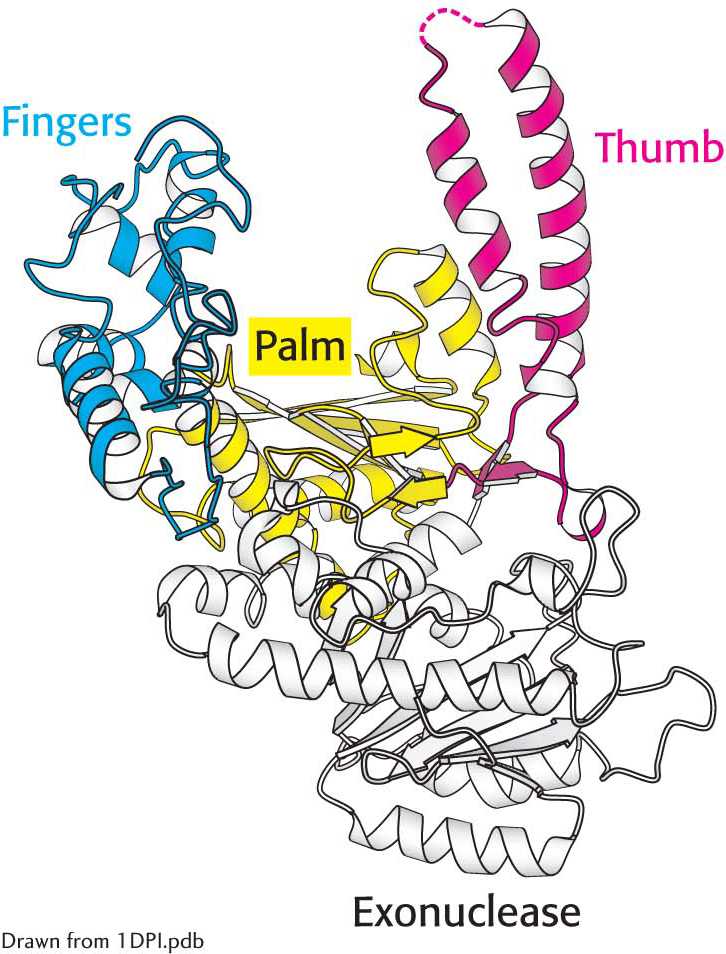
 DNA polymerase structure. The first DNA polymerase structure determined was that of a fragment of E. coli DNA polymerase I called the Klenow fragment. Notice that, like other DNA polymerases, the polymerase unit resembles a right hand with fingers (blue), palm (yellow), and thumb (red). The Klenow fragment also includes an exonuclease domain that removes incorrect nucleotide bases.
DNA polymerase structure. The first DNA polymerase structure determined was that of a fragment of E. coli DNA polymerase I called the Klenow fragment. Notice that, like other DNA polymerases, the polymerase unit resembles a right hand with fingers (blue), palm (yellow), and thumb (red). The Klenow fragment also includes an exonuclease domain that removes incorrect nucleotide bases.
The Specificity of Replication Is Dictated by the Complementarity of Bases
Because DNA is the repository of genetic information, it must be replicated with high accuracy. Each base added to the growing strand should be the Watson–

What is the basis of DNA polymerase’s low error rate? The answer to this question is complex, but one important factor is induced fit—
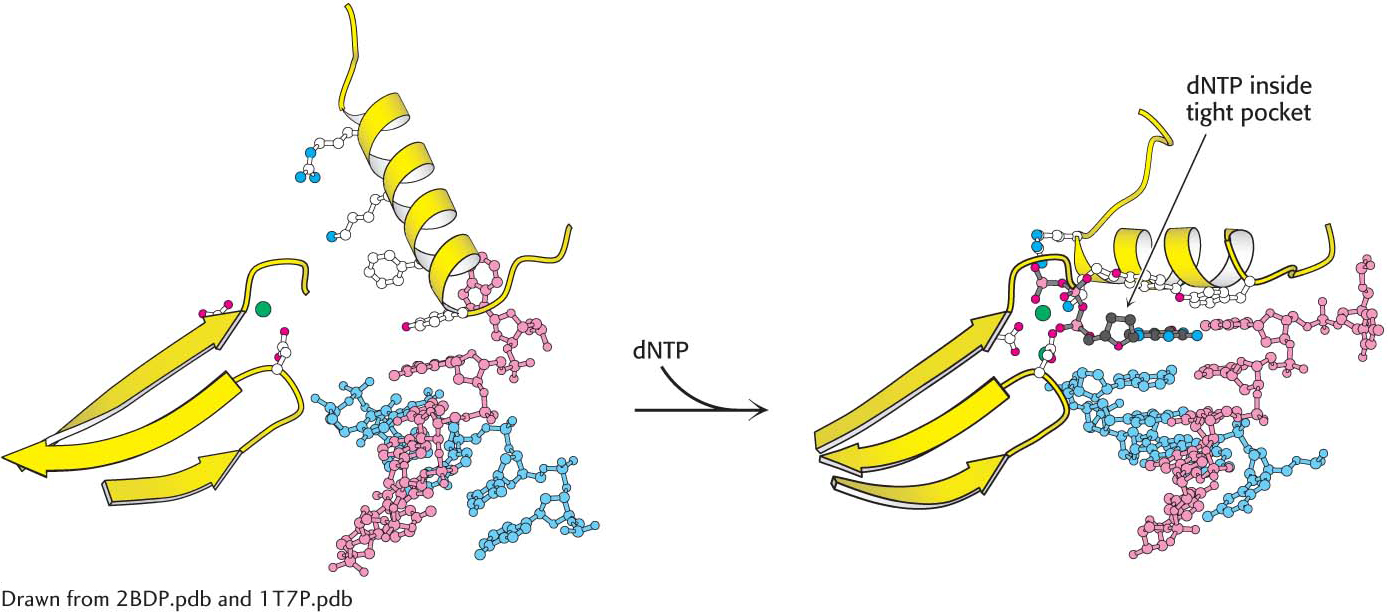

 Shape selectivity. The binding of a deoxyribonucleoside triphosphate (dNTP) to DNA polymerase induces a conformational change, generating a tight pocket for the base pair consisting of the dNTP and its partner on the template strand. Such a conformational change is possible only when the dNTP corresponds to the Watson–
Shape selectivity. The binding of a deoxyribonucleoside triphosphate (dNTP) to DNA polymerase induces a conformational change, generating a tight pocket for the base pair consisting of the dNTP and its partner on the template strand. Such a conformational change is possible only when the dNTP corresponds to the Watson– CLINICAL INSIGHT
CLINICAL INSIGHTThe Separation of DNA Strands Requires Specific Helicases and ATP Hydrolysis
For a double-

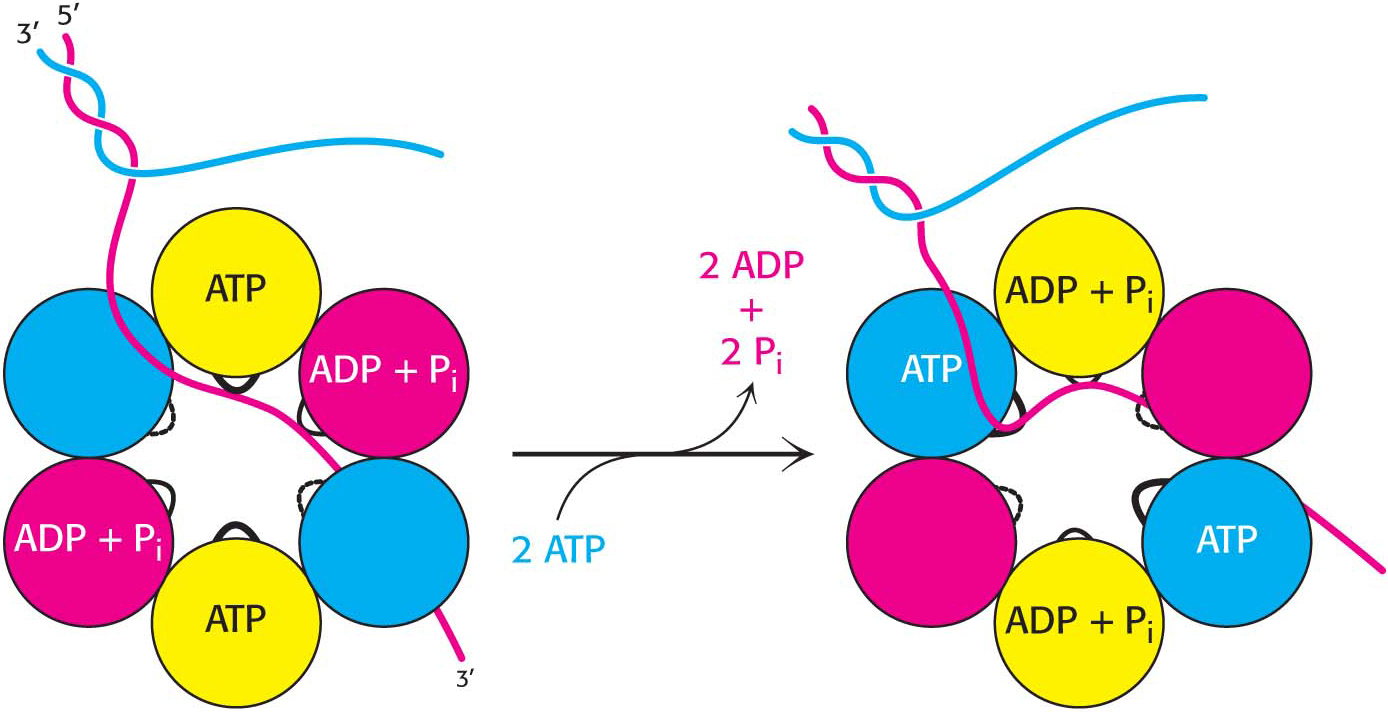
Helicases are a large and diverse family of enzymes taking part in many biological processes. The helicases in DNA replication are typically oligomers containing six subunits that form a ring structure. Each subunit has a loop that extends toward the center of the ring structure and interacts with DNA. A possible mechanism for the action of a helicase is shown in Figure 34.7. Two subunits are bound to ATP, two to ADP and Pi, and two are initially free of nucleotides. One of the strands of the double helix passes through the hole in the center of the helicase, bound to the loops on two adjacent subunits, one of which has bound ATP and the other of which has bound ADP + Pi. The binding of ATP to the subunit that initially had no bound nucleotides results in a conformational change within the entire hexamer, leading to the release of ADP + Pi from two subunits and the binding of the single-
Topoisomerases Prepare the Double Helix for Unwinding
Most naturally occurring DNA molecules are negatively supercoiled. What is the basis for this prevalence? As discussed in Chapter 33, negative supercoiling arises from the unwinding or underwinding of DNA. In essence, negative supercoiling prepares DNA for processes requiring separation of the DNA strands, such as replication. The presence of supercoils in the immediate area of unwinding would, however, make unwinding difficult (Figure 34.8). Therefore, negative supercoils must be continuously removed, and the DNA relaxed, as the double helix unwinds.

DID YOU KNOW?
To gyrate is to move in a circle or spiral or to revolve, usually about a fixed point or on an axis.
Enzymes called topoisomerases introduce or eliminate supercoils by temporarily cleaving DNA. Type I topoisomerases catalyze the relaxation of supercoiled DNA, a thermodynamically favorable process. Type II topoisomerases utilize free energy from ATP hydrolysis to add negative supercoils to DNA. In bacteria, type II topoisomerase is called DNA gyrase.
 CLINICAL INSIGHT
CLINICAL INSIGHTBacterial Topoisomerase Is a Therapeutic Target
DNA gyrase is the target of several antibiotics that inhibit this bacterial topoisomerase much more than the eukaryotic one. Novobiocin blocks the binding of ATP to gyrase. Nalidixic acid and ciprofloxacin, in contrast, interfere with the breakage and rejoining of DNA strands. These two gyrase inhibitors are widely used to treat urinary-
Ciprofloxacin, more commonly known as “cipro,” became a “celebrity” in the United States, owing to the anthrax poisonings in the fall of 2001 (Figure 34.9). It is a potent broad-

Many Polymerases Proofread the Newly Added Bases and Excise Errors
Many polymerases further enhance the fidelity of replication by the use of proofreading mechanisms. One polymerase from E. coli, DNA polymerase I used in DNA replication and repair, displays an exonuclease activity in addition to the polymerase activity. The exonuclease removes mismatched nucleotides from the 3′ end of DNA by hydrolysis. If the wrong nucleotide is inserted, the malformed product is not held as tightly in the polymerase active site. It is likely to flop about because of the weaker hydrogen bonding and to find itself in the exonuclease active site, where the trespassing nucleotide is removed (Figure 34.10). This flopping is the result of Brownian motion.

QUICK QUIZ 1
What are the three key enzymes required for DNA synthesis, and what biochemical challenges to replication do they address?
DNA polymerases, helicases, and topoisomerases. Polymerases faithfully replicate the DNA. Helicases unwind the double helix, allowing the polymerases access to the base sequence. Topoisomerases relax the DNA by removing negative supercoils.
How does the enzyme sense whether a newly added base is correct? First, an incorrect base will not pair correctly with the template strand and will be unlikely to be linked to the new strand. Second, even if an incorrect base is inserted into the new strand, it is likely to be deleted. After the addition of a new nucleotide, the DNA is pulled by one base pair into the enzyme. If an incorrect base is incorporated, the enzyme stalls owing to the structural disruption caused by the presence of a non-