37.1 Eukaryotic Cells Have Three Types of RNA Polymerases

In bacteria, RNA is synthesized by a single kind of polymerase. In contrast, the nucleus of a eukaryote contains three types of RNA polymerase differing in a number of properties, including template specificity and susceptibility to α-amanitin (Table 37.1). α-Amanitin is a toxin produced by the mushroom Amanita phalloides (Figure 37.2) that binds very tightly to RNA polymerase II and thereby blocks the elongation phase of RNA synthesis. Higher concentrations of α-amanitin inhibit polymerase III, whereas polymerase I is insensitive to this toxin. This pattern of sensitivity is highly conserved throughout the animal and plant kingdoms.
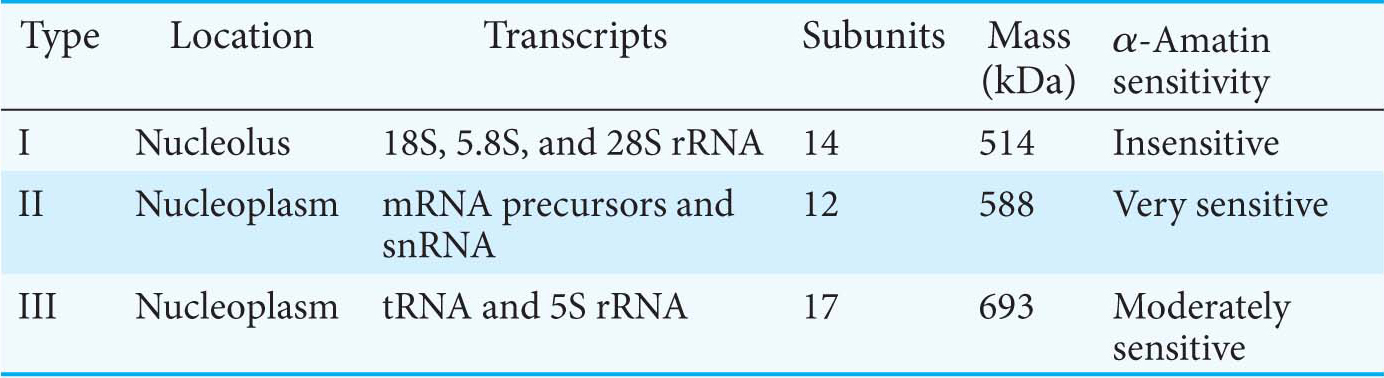
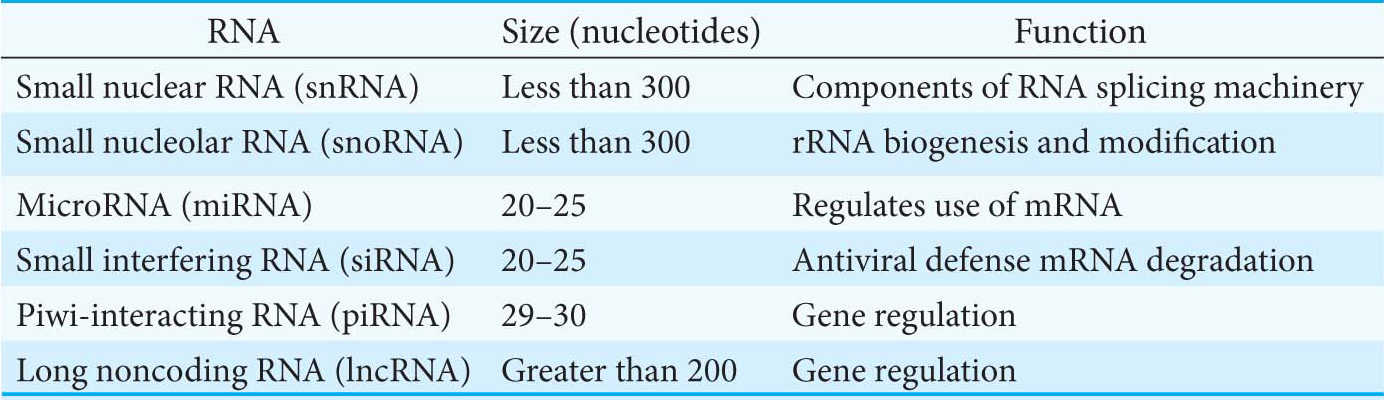
RNA polymerase I is located in nucleoli, where it transcribes the three ribosomal RNAs 18S, 5.8S, and 28S ribosomal RNA as a single transcript. RNA polymerase II is located in the nucleoplasm, the fluid and structural components in the nucleus that surround the chromosomes and nucleoli. RNA polymerase II synthesizes the precursors of messenger RNA as well as several small RNA molecules, such as those of the splicing apparatus. The other ribosomal RNA molecule (5S rRNA, Chapter 39) and all the transfer RNA molecules are synthesized by RNA polymerase III, which also is located in the nucleoplasm. In addition to mRNA, rRNA, and tRNA, there are several other classes of RNA (Table 37.2). The function of some of these RNAs will be considered in Section 16.
Although all eukaryotic RNA polymerases are similar to one another and to bacterial RNA polymerase in structure and function, RNA polymerase II contains a unique carboxyl-
Eukaryotic genes, like bacterial genes, require promoters for transcription initiation. Like bacterial promoters, eukaryotic promoters consist of sequences that serve to attract the polymerases to the start sites. As is the case for bacteria, a promoter and the gene that it regulates are always on the same molecule of DNA. Consequently, promoters are referred to as cis-
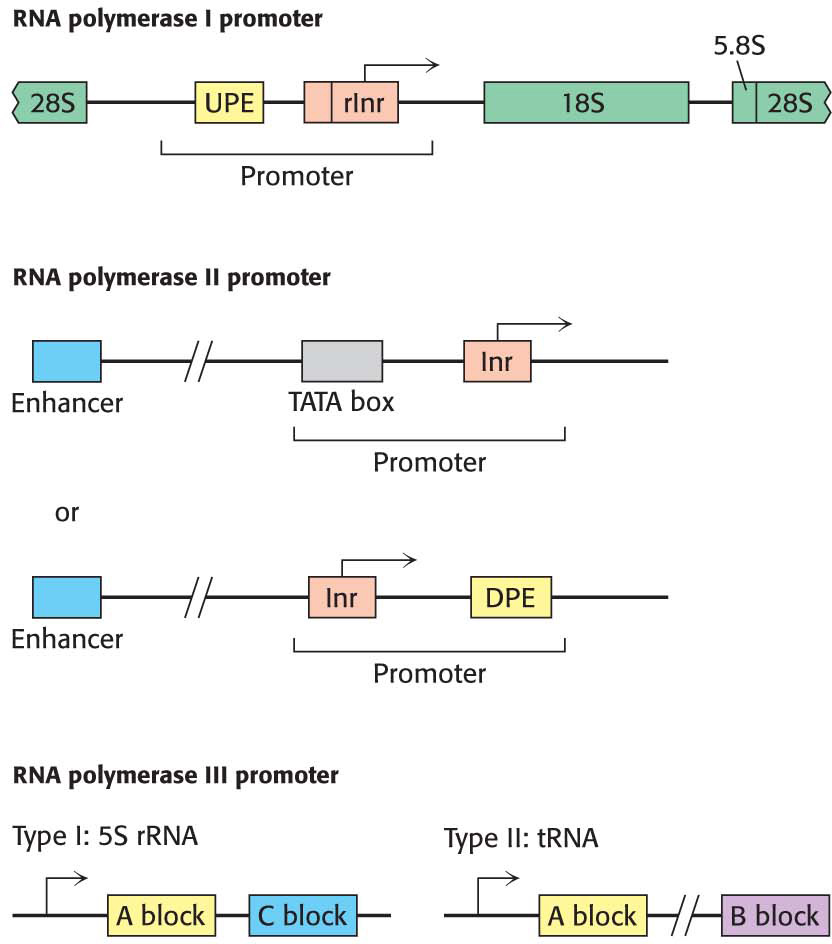
RNA Polymerase I. The ribosomal DNA (rDNA) transcribed by polymerase I is arranged in several hundred tandem repeats, each containing a copy of the three rRNA genes. The promoter sequences are located in stretches of DNA separating the gene repeats. At the transcriptional start site lies a sequence called the ribosomal initiator element (rInr). Farther upstream, 150 to 200 bp from the start site, is the upstream promoter element (UPE). Both elements aid transcription by binding proteins that recruit RNA polymerase I.
RNA Polymerase II. Promoters for RNA polymerase II, like bacterial promoters, include a set of conserved-
sequence elements that define the start site and recruit the polymerase. However, the promoter can contain any combination of possible elements. The diverse array of promoter elements also includes enhancer elements, unique to eukaryotes, that can be very distant (more than 1 kb) from the start site. RNA Polymerase III. Promoters for RNA polymerase III are within the transcribed sequence, downstream of the start site. There are two types of promoters for RNA polymerase III. Type I promoters, found in the 5S rRNA gene, contain two short sequences known as the A block and the C block. Type II promoters, found in tRNA genes, consist of two 11-
bp sequences, the A block and the B block, situated about 15 bp from either end of the gene.
DID YOU KNOW?
Cis-
Trans-