PROBLEMS
Question 38.1
1. Like Jordan and Pippen. Match each term with its description. ✓ 4
Exon Intron Pre- 5′ Cap Poly A tail Splicing Spliceosome snRNA snRNP Alternative splicing | Encodes part of the final mRNA Has a distinctive 5′–5′ linkage Allows one gene to encode several mRNAs Removes introns and joins exons The initial product of RNA polymerase II Catalytic component of the splicing machinery A 3′ posttranscriptional addition A complex that coordinates two transesterification reactions Removed from initial transcript Protein– |
Question 38.2
2. Nice hat. Describe the 5′ cap.
Question 38.3
3. Tales of the end. What are the steps required for the formation of a poly(A) tail on an mRNA molecule?
Question 38.4
4. Maturing. What are the three most common modifications by which primary transcripts are converted into mature mRNA?
Question 38.5
5. A coupling device. What structural component of RNA polymerase II that is lacking in RNA polymerases I and III allows modification of the newly synthesized RNA?
Question 38.6
6. Polymerase inhibition. Cordycepin inhibits poly(A) synthesis at low concentrations and RNA synthesis at higher concentrations.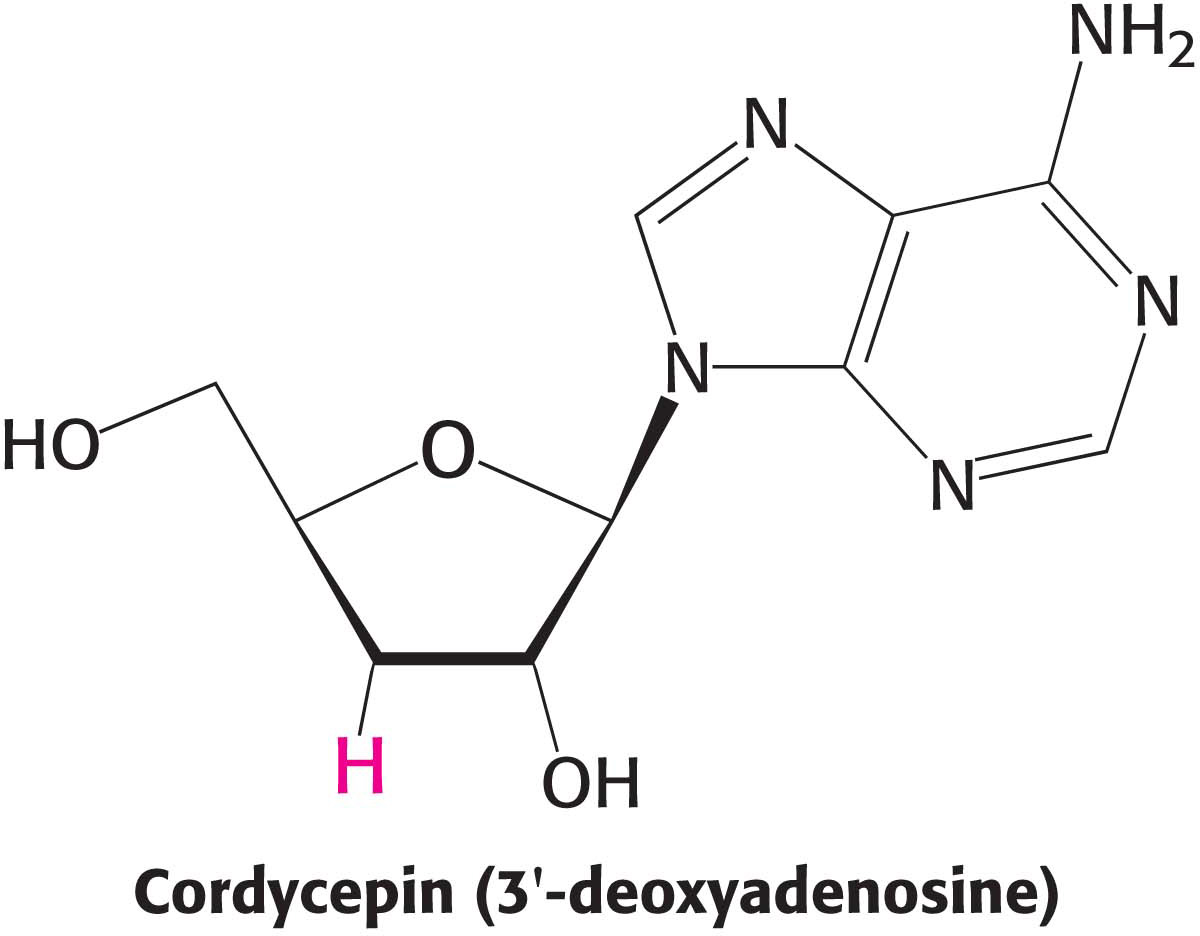
(a) What is the basis of inhibition by cordycepin?
(b) Why is poly(A) synthesis more sensitive to the presence of cordycepin?
(c) Does cordycepin need to be modified to exert its effect?
Question 38.7
7. Teamwork. What is a spliceosome, and of what is it composed?
Question 38.8
8. Recruiting the right team. What is the role of the carboxyl-
Question 38.9
9. From a few, many. Explain why alternative splicing effectively enlarges the size of the genome.
Question 38.10
10. Alternative splicing. A gene contains eight sites where alternative splicing is possible. With the assumption that the splicing pattern at each site is independent of those at all other sites, how many splicing products are possible? ✓ 4
Question 38.11
11. A good idea at the time. George Beadle and Edward Tatum were awarded the Nobel Prize for their research on the relation of genes to enzymes. In a paper published in 1941, they proposed the “one gene, one enzyme hypothesis,” which postulated that each gene encodes a single enzyme or, more generally, a single protein. Although this hypothesis was very influential, we now know that it is an oversimplification. Explain why.
Question 38.12
12. Searching for similarities. Compare self-
Chapter Integration Problems
Question 38.13
13. Proteome complexity. What process considered in this chapter makes the proteome more complex than the genome? What processes might further enhance this complexity? ✓ 4
Question 38.14
14. Separation technique. Suggest a means by which you could separate mRNA from the other types of RNA in a eukaryotic cell.
Question 38.15
15. A cell-
Challenge Problems
Question 38.16
16. An extra piece. In one type of mutation leading to a form of thalassemia, the mutation of a single base (G → A) generates a new 3′ splice site (blue in the adjoining illustration) like the normal one (yellow) but farther upstream.
What is the amino acid sequence of the extra segment of protein synthesized in a thalassemic patient having a mutation leading to aberrant splicing? The reading frame after the splice site begins with TCT.
Question 38.17
17. A long-
Selected Readings for this chapter can be found online at www.whfreeman.com/