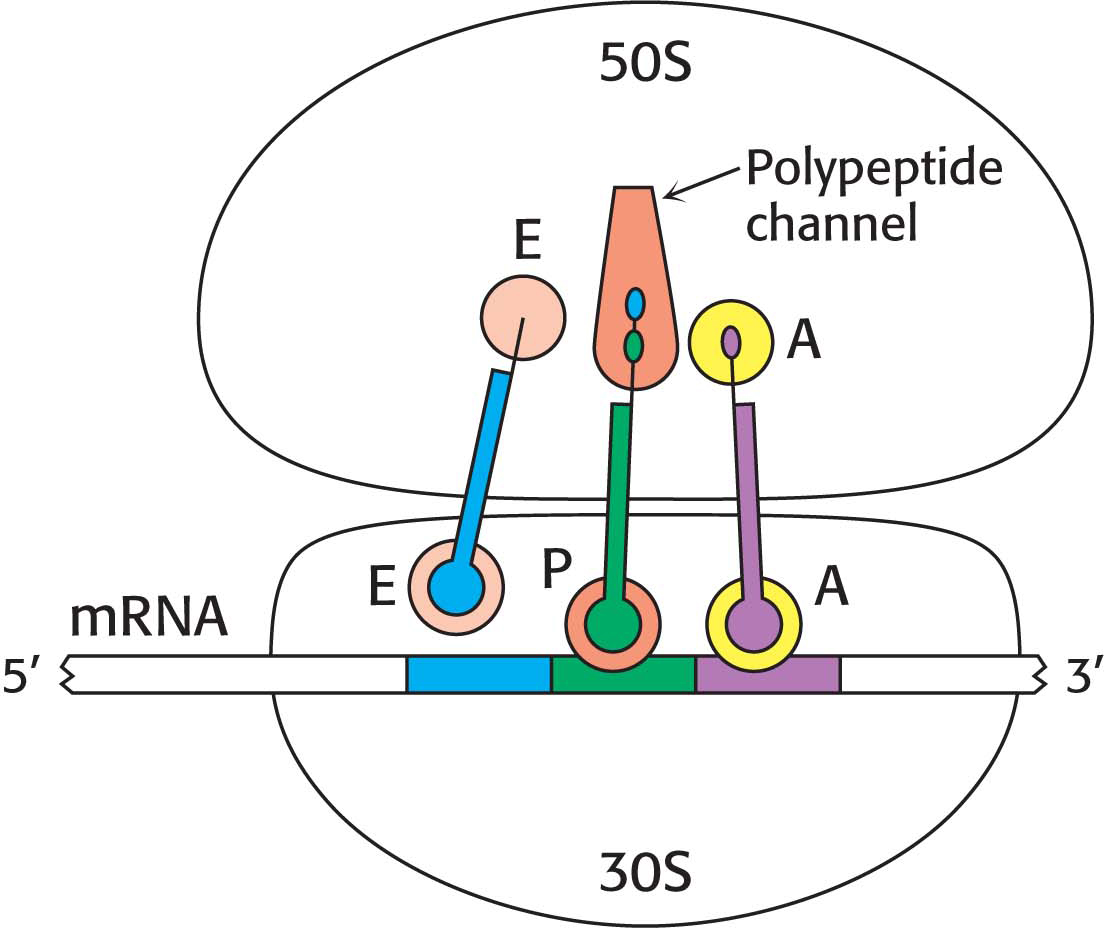
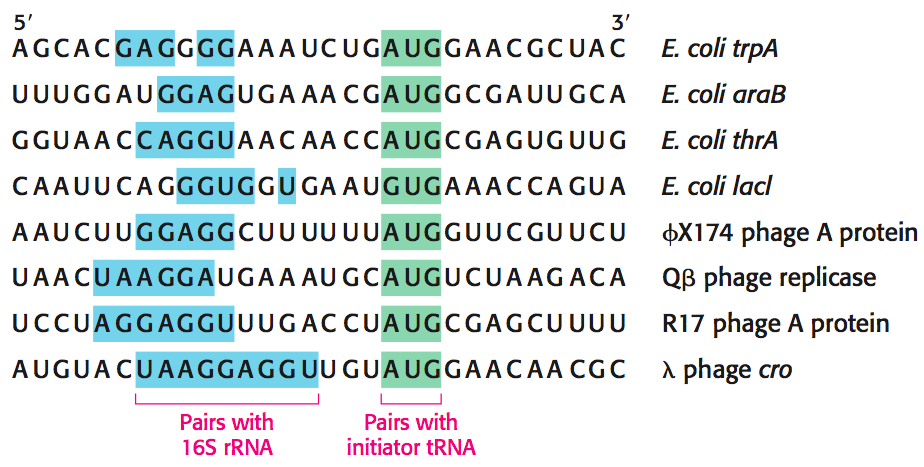Ribosomes Have Three tRNA-Binding Sites That Bridge the 30S and 50S Subunits
The three tRNA-binding sites on ribosomes are arranged to allow the sequential formation of peptide bonds between amino acids encoded by the codons on mRNA (Figure 40.1). The mRNA fragment being translated at a given moment is bound within the 30S subunit. Each of the tRNA molecules is in contact with both the 30S subunit and the 50S subunit. At the 30S end, two of the three tRNA molecules are bound to the mRNA through anticodon–codon base pairs. These binding sites are called the A site (for aminoacyl) and the P site (for peptidyl). The third tRNA molecule is bound to an adjacent site called the E site (for exit).
The other end of each tRNA molecule interacts with the 50S subunit. The acceptor stems of the tRNA molecules occupying the A site and the P site converge at a site where a peptide bond is formed. A tunnel connects this site to the back of the ribosome, through which the polypeptide chain passes during synthesis (Figure 40.2).
The Start Signal Is AUG Preceded by Several Bases That Pair with 16S Ribosomal RNA
How does protein synthesis start? In other words, how are the codons appropriately positioned in the A and P sites? The simplest possibility would be for the first three nucleotides of each mRNA to serve as the first codon; no special start signal would then be needed. However, the experimental fact is that translation in bacteria does not begin immediately at the 5′ terminus of mRNA. Indeed, the first translated codon is nearly always more than 25 nucleotides away from the 5′ end. Furthermore, in bacteria, many mRNA molecules are polycistronic—that is, they encode two or more polypeptide chains. Recall that, in the lac operon, the mRNA product encodes three enzymes. Each of these three proteins has its own start and stop signals on mRNA. In fact, all known mRNA molecules contain signals that define the beginning and the end of each encoded polypeptide chain.
Page 723
A clue to the mechanism of initiation was the finding that nearly half the amino-terminal residues of proteins in E. coli are methionine. In fact, the initiating codon in mRNA is usually AUG (methionine). However, the initiating codon alone is not up to the task of protein-synthesis initiation. In addition, each initiator region contains a purine-rich sequence, called the Shine–Dalgarno sequence (named after John Shine and Lynn Dalgarno, who first described the sequence), centered about 10 nucleotides on the 5′ side of the initiator codon (Figure 40.3). Nucleotide sequences in mRNA, such as the Shine–Dalgarno sequence, that are not translated are called untranslated regions (UTRs) and may be at the 5′ end of the mRNA (5′-UTR) or at the 3′ end (3′-UTR). Untranslated regions usually function to regulate the usage of mRNA molecules.
The Shine–Dalgarno sequence interacts with a complementary sequence on the 3′ end of the 16S rRNA component of the ribosome’s small subunit. Thus, two kinds of interactions determine where protein synthesis starts in bacteria: (1) the pairing of mRNA bases with the 3′ end of 16S rRNA and (2) the pairing of the initiator codon on mRNA with the anticodon of an initiator tRNA molecule.
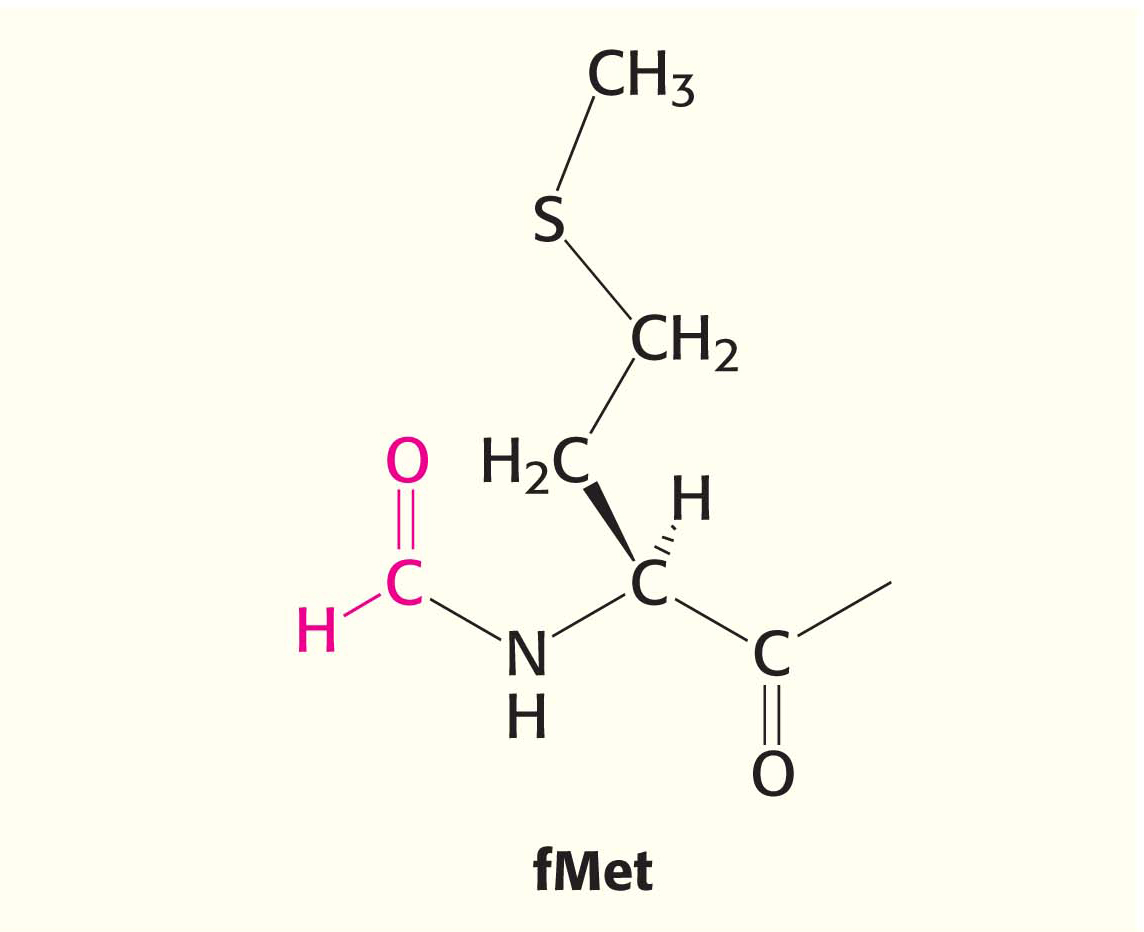
Bacterial Protein Synthesis Is Initiated by Formylmethionyl Transfer RNA
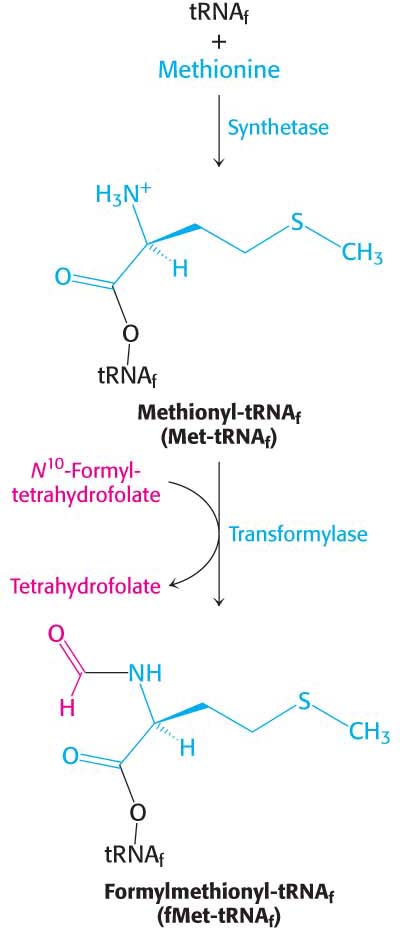
As stated earlier, methionine is the first amino acid in many E. coli proteins. However, the methionine residue found at the amino-terminal end of E. coli proteins is usually modified. In fact, protein synthesis in bacteria starts with the modified amino acid N-formylmethionine (fMet). A special tRNA brings formylmethionine to the ribosome to initiate protein synthesis. This initiator tRNA (abbreviated as tRNAf) differs from the tRNA that inserts methionine in internal positions (abbreviated as tRNAm). The subscript “f” indicates that methionine attached to the initiator tRNA can be formylated, whereas it cannot be formylated when attached to tRNAm. Although virtually all proteins synthesized in E. coli begin with formylmethionine, in approximately one-half of the proteins, N-formylmethionine is removed when the nascent chain is 10 amino acids long.
Methionine is linked to these two kinds of tRNAs by the same aminoacyl-tRNA synthetase. A specific enzyme then formylates the amino group of the methionine molecule that is attached to tRNAf (Figure 40.4). The activated formyl donor in this reaction is N10-formyltetrahydrofolate, a folate derivative that carries activated one-carbon units. Free methionine and methionyl-tRNAm are not substrates for this transformylase.
Page 724
Formylmethionyl-tRNAf Is Placed in the P Site of the Ribosome in the Formation of the 70S Initiation Complex
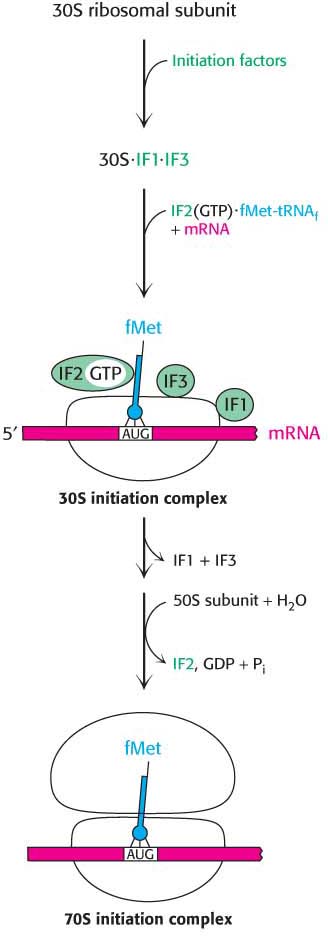
Messenger RNA and formylmethionyl-tRNAf must be brought to the ribosome for protein synthesis to begin. How is this task accomplished? Three protein initiation factors (IF1, IF2, and IF3) are essential. The 30S ribosomal subunit first forms a complex with IF1 and IF3 (Figure 40.5). The binding of these factors to the 30S subunit prevents it from prematurely joining the 50S subunit to form a dead-end 70S complex, devoid of mRNA and fMet-tRNAf. Initiation factor 2, a GTPase, binds GTP, and the concomitant conformational change enables IF2 to associate with formylmethionyl-tRNAf. The IF2–GTP–initiator-tRNA complex binds with mRNA (correctly positioned by the interaction of the Shine–Dalgarno sequence with the 16S rRNA) and the 30S subunit to form the 30S initiation complex. Structural changes then lead to the ejection of IF1 and IF3. IF2 stimulates the association of the 50S subunit to the complex. The GTP bound to IF2 is hydrolyzed upon arrival of the 50S subunit, releasing IF2. The result is a 70S initiation complex. The formation of the 70S initiation complex is the rate-limiting step in protein synthesis.
When the 70S initiation complex has been formed, the ribosome is ready for the elongation phase of protein synthesis. The fMet-tRNAf molecule occupies the P site on the ribosome, positioned so that its anticodon pairs with the initiating codon on mRNA. The other two sites for tRNA molecules, the A site and the E site, are empty. This interaction establishes the reading frame for the translation of the entire mRNA. After the initiator codon has been located, groups of three nonoverlapping nucleotides are defined and subsequently translated as protein synthesis takes place.
Elongation Factors Deliver Aminoacyl-tRNA to the Ribosome
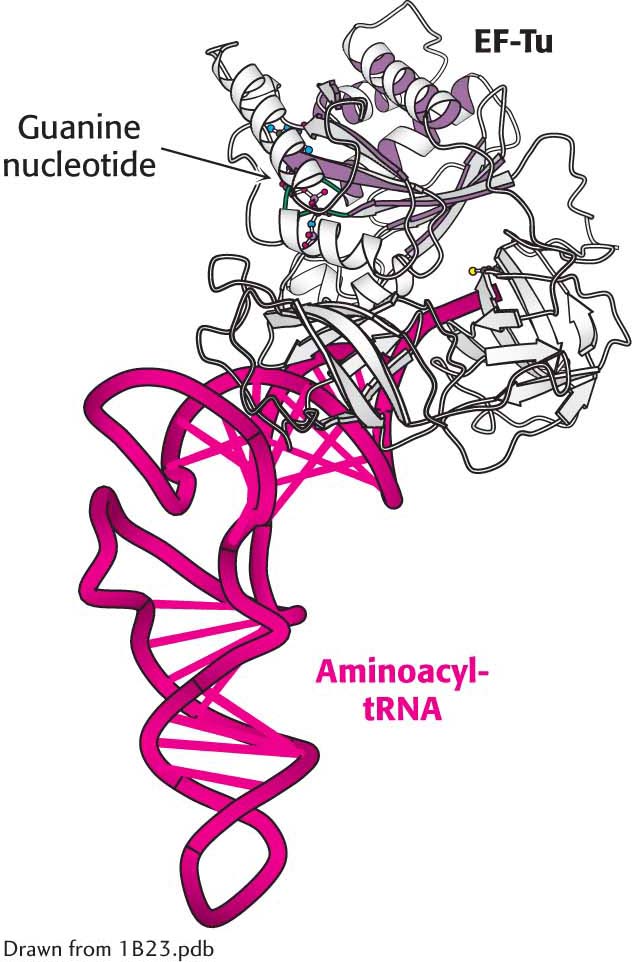
At this point, fMet-tRNAf occupies the P site, and the A site is vacant, as stated. The particular species inserted into the empty A site depends on the mRNA codon in the A site. However, the appropriate aminoacyl-tRNA does not simply leave the synthetase and diffuse to the A site. Rather, it is delivered to the A site in association with a 43-kDa protein called elongation factor Tu (EF-Tu), which requires GTP for activity. Elongation factor Tu, the most abundant bacterial protein, binds aminoacyl-tRNA only in the GTP form (Figure 40.6). The binding of EF-Tu to aminoacyl-tRNA serves two functions. First, EF-Tu protects the delicate ester linkage in aminoacyl-tRNA from hydrolysis. Second, EF-Tu contributes to the accuracy of protein synthesis because GTP hydrolysis and expulsion of the EF-Tu-GDP complex from the ribosome occur only if the pairing between the anticodon and the codon is correct. EF-Tu interacts with the 16S RNA. Correct codon recognition induces structural changes in the 30S subunit that activates the GTPase activity of EF-Tu, releasing EF-Tu-GDP from the ribosome. These same structural changes also rotate the aminoacyl-tRNA in the A site so that the amino acid is brought into proximity with the aminoacyl-tRNA in the P site, a process called accommodation. Accommodation aligns the amino acids for peptide bond formation.
Page 725
Released EF-Tu is then reset to its GTP form by a second elongation factor, elongation factor Ts. EF-Ts induces the dissociation of GDP. GTP binds to EF-Tu, and EF-Ts is concomitantly released. It is noteworthy that EF-Tu does not interact with fMet-tRNAf. Hence, this initiator tRNA is not delivered to the A site. In contrast, Met-tRNAm, like all other aminoacyl-tRNAs, does bind to EF-Tu. These findings account for the fact that internal AUG codons are not read by the initiator tRNA. Conversely, IF2 recognizes fMet-tRNAf but no other tRNA. The cycle of elongation continues until a termination codon is met.

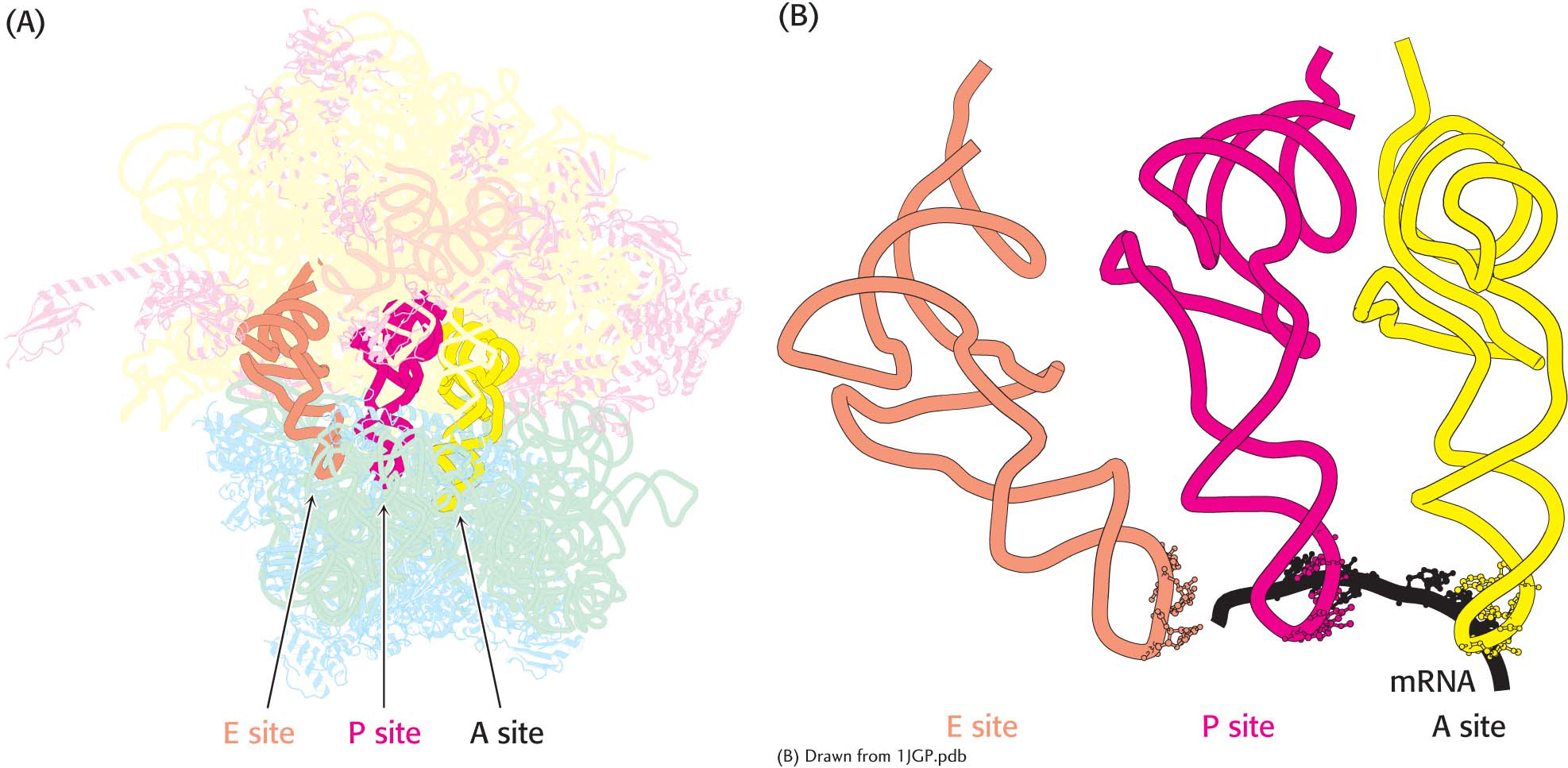
 Binding sites for transfer RNA. (A) Three tRNA molecules are shown in the tRNA-
Binding sites for transfer RNA. (A) Three tRNA molecules are shown in the tRNA-