
41.1 Nucleic Acids Can Be Synthesized from Protein-Sequence Data
In Chapter 5, we purified the receptor by using monoclonal antibodies. What is the next step in characterizing the receptor? There are many possible answers to the question, but an especially common one is to isolate the DNA that encodes the receptor. There are “libraries” of DNA sequences that we can search for sequences corresponding to the receptor. For instance, a complementary DNA (cDNA) library is a collection of DNA sequences representing all of the mRNA expressed by a cell. The enzyme reverse transcriptase uses mRNA as a template to make a DNA copy, or cDNA. If we can isolate the cDNA for the estrogen receptor, we can insert the DNA into bacteria, and the bacteria will produce the estrogen receptor for our experiments. How can we find the estrogen receptor in the cDNA library? We can use our purified estrogen receptor to generate a probe that will allow us to identify the DNA sequences in the library that correspond to the estrogen receptor.
Protein Sequence Is a Guide to Nucleic Acid Information
Chapter 5 examined the techniques used to sequence an entire protein. To search DNA libraries, we need a probe—
Using purified receptor as starting material, we can use mass spectroscopy to determine the amino acid sequence of a part of the receptor. With this information and a copy of the genetic code in hand, we can convert the protein-
DNA Probes Can Be Synthesized by Automated Methods
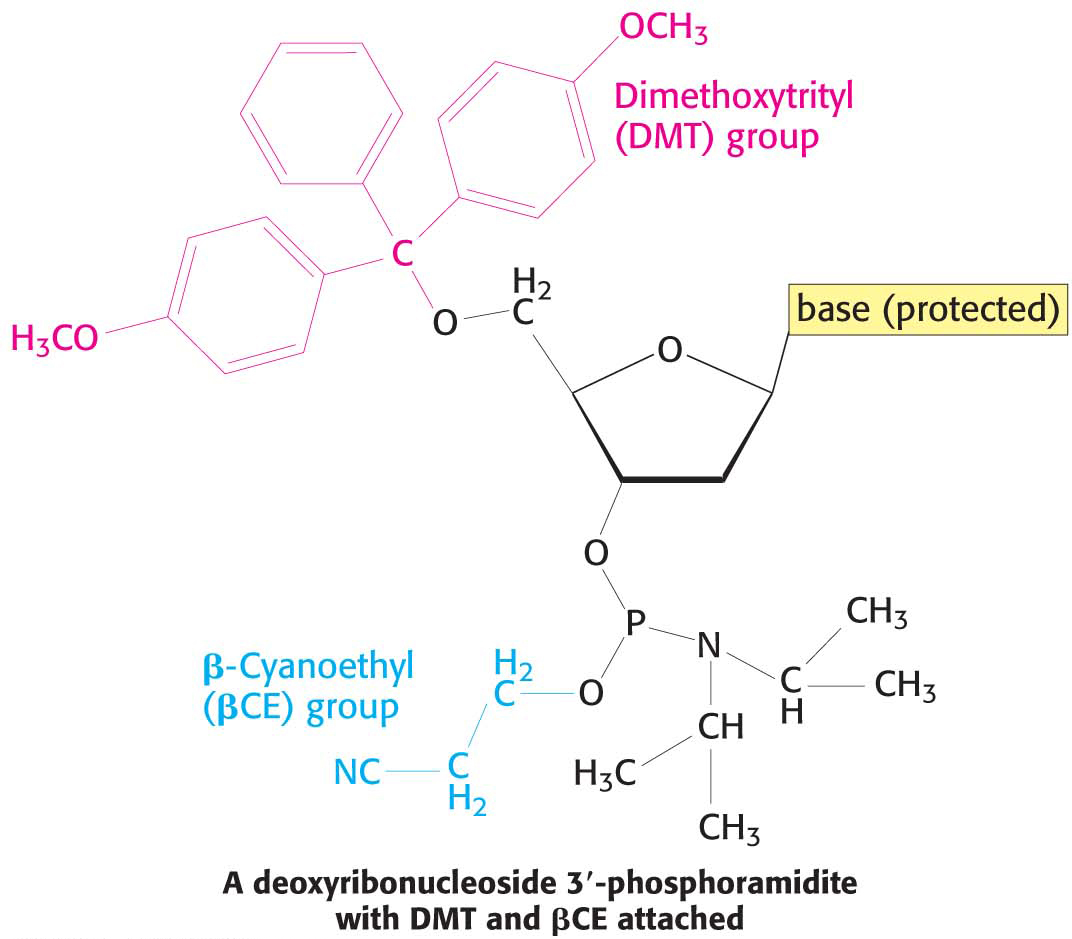
A DNA strand can be synthesized by the sequential addition of activated monomers to a growing strand that is linked to an insoluble support. The activated monomers are protonated deoxyribonucleoside 3′-phosphoramidites in which all but one of the reactive groups of the nucleotide are protected by the temporary attachment of nonreactive groups.
In step 1, the 3′-phosphorus atom of this incoming unit joins the growing strand to form a phosphite triester (Figure 41.1). In step 2, the phosphite triester is oxidized by iodine to stabilize the new linkage. In step 3, the protecting group on the 5′-OH group of the growing strand is removed by the addition of dichloroacetic acid. The DNA strand is now elongated by one unit and ready for another cycle of addition. Each cycle only takes a few minutes and usually elongates more than 99% of the strands.
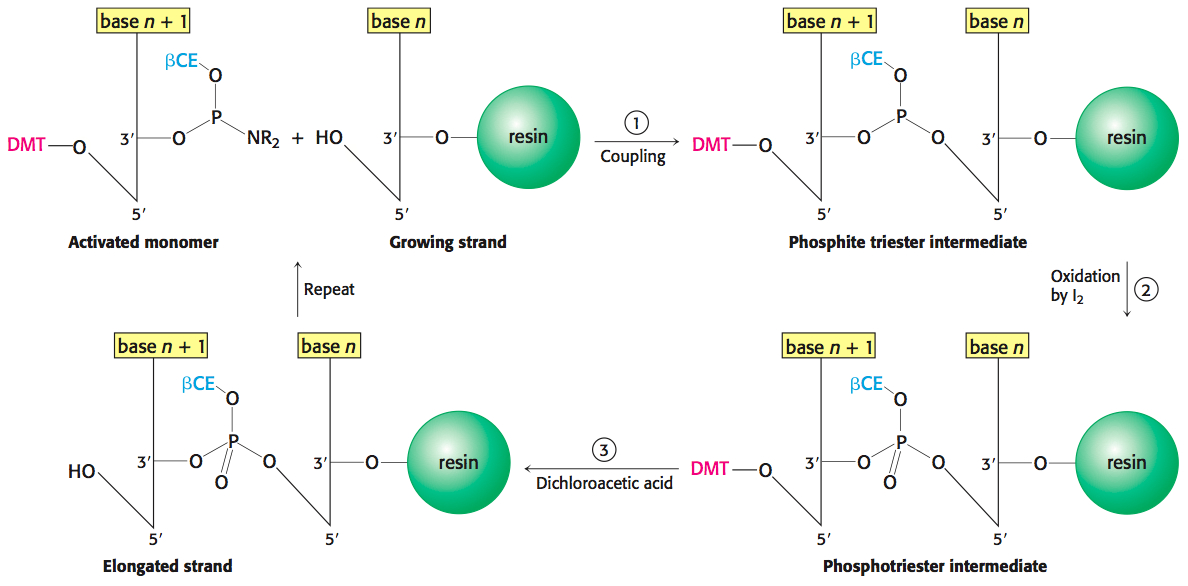
The ability to rapidly synthesize DNA strands of any selected sequence opens many experimental avenues. For example, a synthesized oligonucleotide labeled at one end with 32P or a fluorescent tag can be used to search for a complementary sequence in a collection of DNA sequences. In our experiment, we will make a radioactive DNA probe corresponding to a part of the estrogen receptor’s primary structure. We will use this probe to screen collections of DNA sequences that encode the estrogen receptor. However, before we do so, we must examine the practical characteristics of the DNA that we are looking for and, in the process, we will examine some of the tools for recombinant DNA technology.