PROBLEMS
Question 5.1
1. Knowing that it’s there. Why is an assay necessary to purify a protein? ✓ 4
Question 5.2
2. Pair bonding. Match the terms with the descriptions. ✓ 4
Assay Molecular exclusion chromatography Ion- Affinity chromatography High- Isoelectric focusing Sedimentation coefficient Antigenic determinant (epitope) Monoclonal antibodies Western blotting | Specific site recognized by an antibody Separating proteins on the basis of size differences Based on attraction to a specific chemical group or molecule Allows high resolution and rapid separation Produced by hybridoma cells Separating proteins on the basis of net charge A measure of the rate of movement due to centrifugal force An immunoassay technique preceded by gel electrophoresis Based on the fact that proteins have a pH at which the net charge is zero A means of identifying a protein based on a unique property of the protein |
Question 5.3
3. Salting out. Why do proteins precipitate at high salt concentrations? ✓ 4
Question 5.4
4. Salting in. Although many proteins precipitate at high salt concentrations, some proteins require salt to dissolve in water. Explain why some proteins require salt to dissolve.
Question 5.5
5. Competition for water. What types of R groups would compete with salt ions for water of solvation?
Question 5.6
6. Column choice.
(a) The octapeptide AVGWRVKS was digested with the enzyme trypsin. Would ion-
(b) Suppose that the peptide had, instead, been digested with chymotrypsin. What would be the optimal separation technique? Explain.
Question 5.7
7. Frequently used in shampoos. The detergent sodium dodecyl sulfate (SDS) denatures proteins. Suggest how SDS destroys protein structure.
Question 5.8
8. Making more enzyme? In the course of purifying an enzyme, a researcher performs a purification step that results in an increase in the total activity to a value greater than that present in the original crude extract. Explain how the amount of total activity might increase. ✓ 4
Question 5.9
9. Protein-
|
Purification procedure |
Total protein (mg) |
Total activity (units) |
Specific activity (units mg−1) |
Purification level |
Yield (%) |
|---|---|---|---|---|---|
|
Crude extract |
20,000 |
4,000,000 |
|
1 |
100 |
|
(NH4)2SO4 precipitation |
5000 |
3,000,000 |
|
|
|
|
DEAE– |
1500 |
1,000,000 |
|
|
|
|
Gel- |
500 |
750,000 |
|
|
|
|
Affinity chromatography |
45 |
675,000 |
|
|
|
|
Note: DEAE (diethylaminoethyl) bears a positive charge, and so DEAE- |
|||||
Question 5.10
10. Charge to mass.
(a) Proteins treated with a sulfhydryl reagent such as β-mercaptoethanol and dissolved in sodium dodecyl sulfate have the same charge-
(b) Under what conditions might the statement in part a be incorrect?
(c) Some proteins migrate anomalously in SDS–
Question 5.11
11. Push back. During ultracentrifugation, the force opposing centrifugation is the buoyant force, equal to  . Using the equation for buoyant force, show why a dense particle moves more rapidly than does a less dense one in ultracentrifugation.
. Using the equation for buoyant force, show why a dense particle moves more rapidly than does a less dense one in ultracentrifugation.
Question 5.12
12. A special attraction. What unique property of the estrogen receptor allows for its identification and purification?✓ 5
Question 5.13
13. Many or one. Differentiate between polyclonal and monoclonal antibodies. ✓ 5
Question 5.14
14. A falling out. Explain how immunoprecipitation can be used to purify proteins. ✓ 5
Question 5.15
15. Don’t you mean who? What is an ELISA, and how is it used? ✓ 5
Question 5.16
16. John Wayne’s favorite. Describe western blotting. ✓ 5
Question 5.17
17. A question of efficiency. The Edman method of protein sequencing can be used to determine a sequence of proteins no longer than approximately 50 amino acids. Why is this length limitation the case? ✓ 5
Question 5.18
18. Divide and conquer. The determination of the mass of a protein by mass spectrometry often does not allow its unique identification among possible proteins within a complete proteome, but determination of the masses of all fragments produced by digestion with trypsin almost always allows unique identification. Explain.
Chapter Integration Problem
Question 5.19
19. Quaternary structure. A protein was purified to homogeneity. Determination of the mass by gel-
Data Interpretation Problems
Question 5.20
20. Protein solubility. The isoelectric point (pI) of a protein is the pH at which a protein has no net charge. The pI is also the pH at which a protein is least soluble. The graph below shows the solubility of a protein as a function of pH in solutions of different ionic strengths. ✓ 4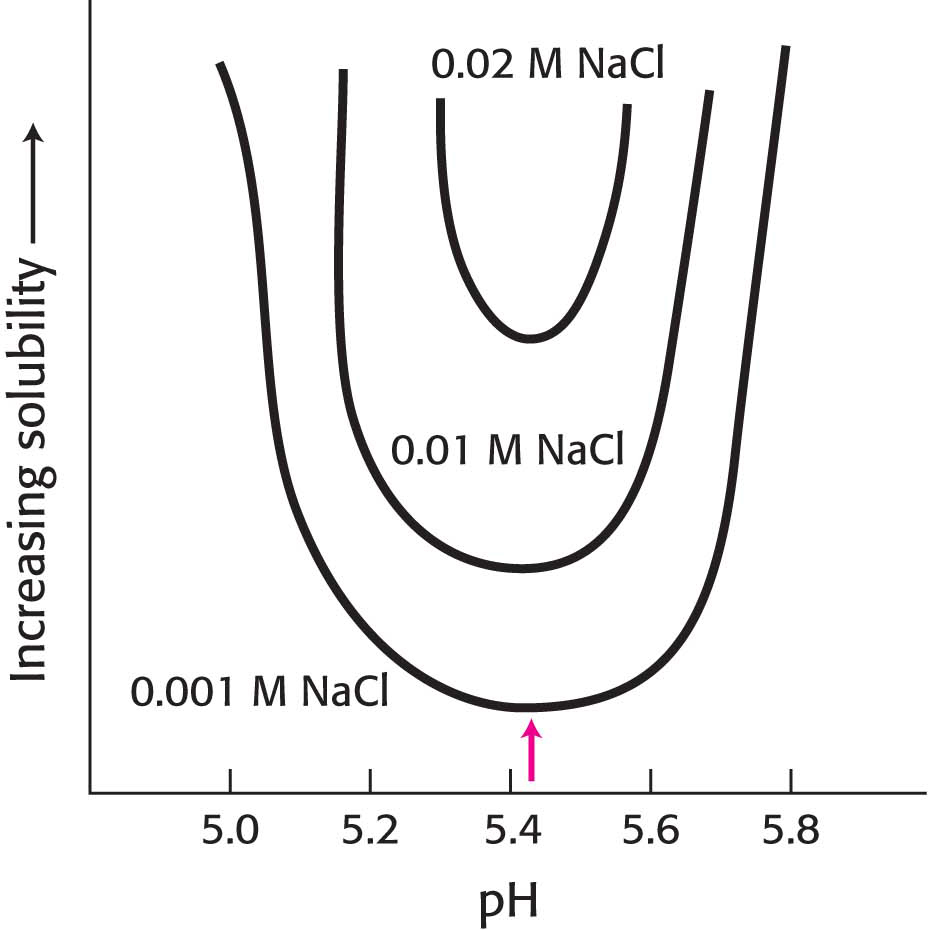
(a) Why is the protein least soluble at its pI (indicated by the red arrow in the figure)?
(b) Why does increasing the salt concentration increase the protein’s solubility?
Question 5.21
21. Protein sequencing 1. Determine the sequence of hexapeptide on the basis of the following data. Note: When the sequence is not known, a comma separates the amino acids. (Table 5.3)
Amino acid composition: (2R,A,S,V,Y)
Amino-
Trypsin digestion: (R,A,V) and (R,S,Y)
Carboxypeptidase A digestion: no digestion
Chymotrypsin digestion: (A,R,V,Y) and (R,S)
Question 5.22
22. Protein sequencing 2. Determine the sequence of a peptide consisting of 14 amino acids on the basis of the following data.
Amino acid composition: (4S,2L,F,G,I,K,M,T,W,Y)
Amino-
Carboxypeptidase A digestion: L
Trypsin digestion: (3S,2L,F,I,M,T,W) (G,K,S,Y)
Chymotrypsin digestion: (F,I,S) (G,K,L) (L,S) (M,T) (S,W) (S,Y)
Amino-
Cyanogen bromide treatment: (2S,F,G,I,K,L,M*,T,Y) (2S,L,W)
M*, methionine detected as homoserine
Challenge Problems
Question 5.23
23. Dialysis. Suppose that you precipitate a protein with 1 M (NH4)2SO4, and you wish to reduce the concentration of the (NH4)2SO4. You take 1 ml of your sample and dialyze it in 1000 ml of buffer. At the end of dialysis, what is the concentration of (NH4)2SO4 in your sample? How could you further lower the (NH4)2SO4 concentration?
Question 5.24
24. Sedimenting spheres. What is the dependence of the sedimentation coefficient s of a spherical protein on its mass? How much more rapidly does an 80-
Selected Readings for this chapter can be found online at www.whfreeman.com/