- Step 1
- Step 2
- Step 3
- Step 4
- Step 5
- Step 6
- Step 7
- Step 8
Chapter 5. Chapter 5: Mapping Bacterial Genes Using Interrupted Mating and Recombination During Bacterial Conjugation
Introduction
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
An Hfr is:
| A. |
| B. |
| C. |
| D. |
| E. |
What differentiates F+ E. coli cells from the Hfr variety? Answer true or false:
Hfr cells contain the fertility factor as a plasmid, while F+ cells do not.
Hfr cells contain the fertility factor integrated into the bacterial chromosome, while F+ cells contain it as a plasmid.
Both Hfr and F+ cells readily convert F- cells into fertility positive cells.
Only F+ cells readily convert F- cells into fertility positive cells.
What role does an Hfr cell play during conjugation? Answer true or false:
An Hfr cell is a donor of genetic material.
An Hfr cell is a recipient of genetic material.
An Hfr cell always readily transfers a fertility factor to the recipient.
An Hfr cell transfer of genetic information can lead to recombination in the donor.
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
What is interrupted mating?
| A. |
| B. |
| C. |
| D. |
The objective of this problem is mapping bacterial genes. The natural phenomenon that is being exploited is bacterial conjugation. How is conjugation related to mating?
Conjugation is the process by which bacterial cells mate.
Conjugation is one of three ways that bacterial cells mate.
How might interrupting conjugation be useful in generating data that could be analyzed to map bacterial gene order?
At different time points of interruption, the profile of donor genes found within the recipient cells may be different.
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
What is the significance of selecting for the terminally transferred arg+ marker?
Note that this question is at the crux of the analysis. Think carefully about each answer choice below.
| A. |
| B. |
| C. |
| D. |
| E. |
The problem tells us that the arg+ marker is the last one to enter the recipient cell. Knowing this and that our goal is to map the bacterial genes, why select for the arg+ marker?
Knowing that the arg+ marker is the last to enter the donor cell guarantees that all of the other markers are also present in the donor cell.
It is possible to map all of the bacterial genes of interest even if not all are known to be have been transferred to the donor cell.
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
1.
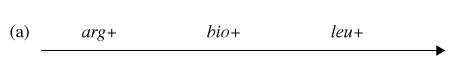
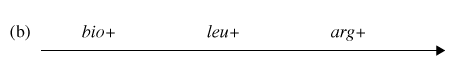
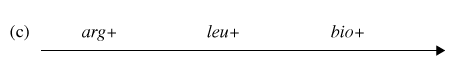
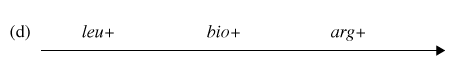
| A. |
| B. |
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
What is the maximum number of crossover events (recombinations) that can occur along a paired homologous region of DNA?
| A. |
| B. |
| C. |
| D. |
| E. |
Can a single crossover produce a viable recombinant (see Figure 5-16)?
Does the length of the homologous sections of DNA make any difference in determining the number of crossover events?
The probability of a crossover occurring is directly proportional to the length of homologously-pairing DNA.
The probability of the number of crossovers is determined by the product rule.
The probability of the number of crossovers is determined by the sum rule.
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
Which of the following statements is true with respect to creating a viable recombinant exconjugant?
| A. |
| B. |
| C. |
| D. |
Can a single crossover produce a viable recombinant (see Figure 5-16)?
What is the difference between the DNA in the recipient cell and the homologous DNA that has entered via bacterial conjugation?
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
You have determined (in step #4) that the correct order of gene transfer from Hfr donor during conjugation is one of the following:
Gene order #1: 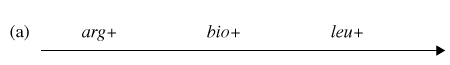
Gene order #2: 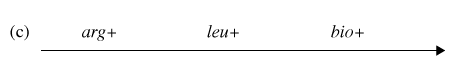
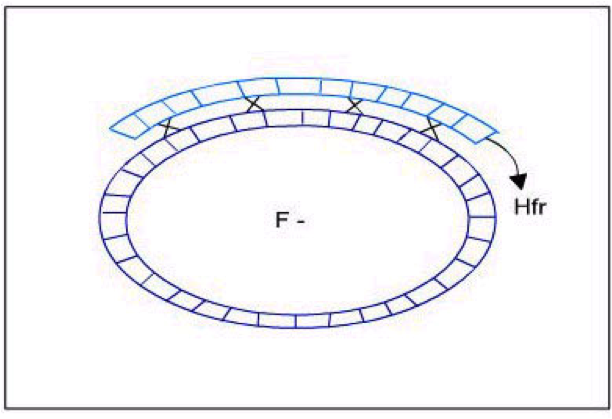
Using the possible gene order information and the diagram depicting possible crossovers, determine the correct gene order.
| A. |
| B. |
| C. |
Now it is time to synthesize the information from the previous six questions. Look at the genotype data provided with this problem, and analyze how each of the four genotypes could form as a result of recombination events within an exconjugant.
A cross is made between two E. coli strains: Hfr arg+ bio+ leu+ × F- arg- bio- leu-. Interrupted mating studies show that arg+ enters the recipient last, and so any arg+ recombinants are selected on a medium containing biotin and leucine only. These recombinants are tested for the presence of bio+ and leu+. The following numbers of individuals are found for each genotype:
arg+ bio+ leu+ 320
arg+ bio+ leu- 8
arg+ bio- leu+ 0
arg+ bio- leu- 48
Unpack the Problem: Break this problem into several parts and arrive at a solution using this guided, step-by-step approach.
- Part A (step 1): Understand what an Hfr strain is and how it functions during conjugation.
- Part B (steps 2-7): What is the gene order?
- Part C (step 8): What are the map distances in recombination percentages?
It should now be possible to determine the map distances in recombination units.
(a) The distance between arg+ and bio+ is:
| A. |
| B. |
| C. |
| D. |
| E. |
| F. |
What is the total number of exconjugants?
How can you use the genotype data provided in this problem to determine the recombination units between selected markers?
Calculate the percent recombinants relative to total exconjugants scored.
Calculate the percent recombinants relative to total exconjugants scored.
General Hints
The map of the three genes is:
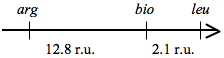
A variety of different double crossover exconjugants can form by different combinations of crossing-over in different regions. The probability of crossing over increases as the distance between selectable markers increases. The distance between selectable markers is determined by calculating the percent recombinants relative to total exconjugants scored. The total number of exconjugants is 320+8+0+48=376. We can determine the recombination units between selected markers using the genotype data provided. Assume a crossover to the left (distal to arg+) of the arg+ marker is constant, and the second crossover can occur in any other region between arg+ and the origin of transfer (arrow of Hfr donor DNA). A crossover between arg+ and bio+ will generate arg+bio-leu- exconjugants. The number of these from the data is: 48/376 = 12.8% or 12.8 recombination units. If the second crossover occurs between bio+ and leu+, then arg+bio+leu- exconjugants are formed (8/376= 2.1% or 2.1 r.u.). If the second crossover occurs between leu+ and the origin of transfer on the Hfr (the arrow in the diagram above), then arg+bio+leu+ exconjugants are formed (320/376 = 85.1% or 85.1 recombination units). This demonstrates that most of the crossovers actually took place proximal to the first selected marker (leu+).