Chapter 2. Molecular Biology I
Agarose Gel Electrophoresis and Bacteria Plating and Treatments
General Purpose
This pre-lab will present some of the general concepts related to some of the techniques used in the fields of microbiology and molecular biology and how these techniques relate to understanding the relationship between DNA, genes, and traits expressed by an organism.
Learning Objectives
General Purpose
- Develop a basic understanding of some of the techniques used in the field of molecular biology.
Conceptual
- Gain an understanding of restriction enzymes and their uses.
- Gain an understanding of plasmids.
Background Information
Deoxyribonucleic acid (DNA) is the most common type of genetic material. The sequence of bases present in the strand of DNA can make up a gene that in turn can confer a given trait on an organism. Differences between organisms, even organisms that are members of the same species, are due in part to differences in the DNA or genes between the organisms.
The field of research commonly known as recombinant DNA technology often uses tools that are found in nature. One of the tools used in molecular biology are bacterial plasmids. Plasmids are small circular DNA molecules that are not a part of the bacterial genome (Figure 11-1).
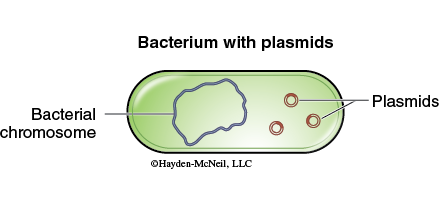
Plasmids usually contain between 1,000 and 100,000 nucleotides. Plasmids can be isolated from one culture of bacterial cells and then transferred to different bacterial cells. This process in which external DNA is transferred to or taken up by an organism is known as transformation. When an organism is transformed, the addition of the genes present on the plasmid DNA changes the genotype of the organism. These new genes may be expressed in the organism resulting in a change in the phenotype of that organism. In this manner traits may be moved from one organism to another.
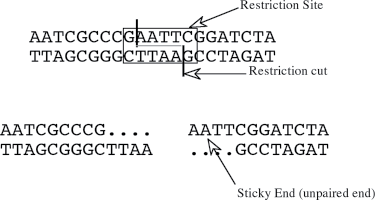
Plasmids may be isolated from bacteria, purified, and the genes present on a bacterial plasmid can be modified by a change in their DNA sequence (mutations). In addition, new genes can be added to or replace the genes on a bacterial plasmid in a process known as gene cloning. Both of these processes usually require that the DNA strands of the plasmid be cut. This is accomplished by using tools from nature. Restriction endonucleases, commonly known as restriction enzymes, cut double-stranded DNA molecules. The substrate specificity of the enzymes results in the cutting of the DNA at specific nucleotide sequences. The sequence at the DNA cut sites is a type of palindrome. A palindrome is a word (or sequence) which reads the same in both directions (radar, madam). In the case of restriction enzymes they cut the DNA at sequences that read the same on both backbones of the DNA. The restriction enzyme EcoRI recognizes a six-base sequence and cuts the DNA backbone within this sequence. The ˇ represents the location of the cut:
Because the cuts occur in both backbones at different locations, the resulting DNA fragments have “sticky ends” (unpaired regions) (Figure 11-2).
The following URL shows an animation of the action of the restriction enzyme EcoRI:
Pre-Lab Quiz
Proceed to the Pre-Lab Quiz