PROBLEMS
Question 4.1
A t instead of an s? Differentiate between a nucleoside and a nucleotide.
Question 4.2
A lovely pair. What is a Watson–
Question 4.3
Chargaff rules! Biochemist Erwin Chargaff was the first to note that, in DNA, [A] = [T] and [G] = [C], equalities now called Chargaff’s rule. Using this rule, determine the percentages of all the bases in DNA that is 20% thymine.
132
Question 4.4
But not always. A single strand of RNA is 20% U. What can you predict about the percentages of the remaining bases?
Question 4.5
Complements. Write the complementary sequence (in the standard 5′ → 3′ notation) for (a) GATCAA, (b) TCGAAC, (c) ACGCGT, and (d) TACCAT.
Question 4.6
Compositional constraint. The composition (in mole-
Question 4.7
Size matters. Why are GC and AT the only base pairs permissible in the double helix?
Question 4.8
Strong, but not strong enough. Why does heat denature, or melt, DNA in solution?
Question 4.9
Uniqueness. The human genome contains 3 billion nucleotides arranged in a vast array of sequences. What is the minimum length of a DNA sequence that will, in all probability, appear only once in the human genome? You need consider only one strand and may assume that all four nucleotides have the same probability of appearance.
Question 4.10
Coming and going. What does it mean to say that the DNA strands in a double helix have opposite directionality or polarity?
Question 4.11
All for one. If the forces—
Question 4.12
Overcharged. DNA in the form of a double helix must be associated with cations, usually Mg2+. Why is this requirement the case?
Question 4.13
Not quite from A to Z. Describe the three forms that a double helix can assume.
Question 4.14
Lost DNA. The DNA of a deletion mutant of λ bacteriophage has a length of 15 μm instead of 17 μm. How many base pairs are missing from this mutant?
Question 4.15
Axial ratio. What is the axial ratio (length:diameter) of a DNA molecule 20 μm long?
Question 4.16
Guide and starting point. Define template and primer as they relate to DNA synthesis.
Question 4.17
An unseen pattern. What result would Meselson and Stahl have obtained if the replication of DNA were conservative (i.e., the parental double helix stayed together)? Give the expected distribution of DNA molecules after 1.0 and 2.0 generations for conservative replication.
Question 4.18
Which way? Explain, on the basis of nucleotide structure, why DNA synthesis proceeds in the 5′-to-
Question 4.19
Tagging DNA.
Suppose that you want to radioactively label DNA but not RNA in dividing and growing bacterial cells. Which radioactive molecule would you add to the culture medium?
Suppose that you want to prepare DNA in which the backbone phosphorus atoms are uniformly labeled with 32P. Which precursors should be added to a solution containing DNA polymerase and primed template DNA? Specify the position of radioactive atoms in these precursors.
Question 4.20
Finding a template. A solution contains DNA polymerase and the Mg2+ salts of dATP, dGTP, dCTP, and TTP. The following DNA molecules are added to aliquots of this solution. Which of them would lead to DNA synthesis?
A single-
stranded closed circle containing 1000 nucleotide units. A double-
stranded closed circle containing 1000 nucleotide pairs. A single-
stranded closed circle of 1000 nucleotides base- paired to a linear strand of 500 nucleotides with a free 3′-OH terminus. A double-
stranded linear molecule of 1000 nucleotide pairs with a free 3′-OH group at each end.
Question 4.21
Retrograde. What is a retrovirus and how does information flow for a retrovirus differ from that for the infected cell?
Question 4.22
The right start. Suppose that you want to assay reverse transcriptase activity. If polyriboadenylate is the template in the assay, what should you use as the primer? Which radioactive nucleotide should you use to follow chain elongation?
Question 4.23
Essential degradation. Reverse transcriptase has ribonuclease activity as well as polymerase activity. What is the role of its ribonuclease activity?
Question 4.24
Virus hunting. You have purified a virus that infects turnip leaves. Treatment of a sample with phenol removes viral proteins. Application of the residual material to scraped leaves results in the formation of progeny virus particles. You infer that the infectious substance is a nucleic acid. Propose a simple and highly sensitive means of determining whether the infectious nucleic acid is DNA or RNA.
Question 4.25
Mutagenic consequences. Spontaneous deamination of cytosine bases in DNA takes place at low but measurable frequency. Cytosine is converted into uracil by loss of its amino group. After this conversion, which base pair occupies this position in each of the daughter strands resulting from one round of replication? Two rounds of replication?
Question 4.26
Information content.
How many different 8-
mer sequences of DNA are there? (Hint: There are 16 possible dinucleotides and 64 possible trinucleotides.) We can quantify the information- carrying capacity of nucleic acids in the following way. Each position can be one of four bases, corresponding to two bits of information (22 = 4). Thus, a chain of 5100 nucleotides corresponds to 2 × 5100 = 10,200 bits, or 1275 bytes (1 byte = 8 bits). 133
How many bits of information are stored in an 8-
mer DNA sequence? In the E. coli genome? In the human genome? Compare each of these values with the amount of information that can be stored on a computer compact disc, or CD (about 700 megabytes).
Question 4.27
Key polymerases. Compare DNA polymerase and RNA polymerase from E. coli in regard to each of the following features: (a) activated precursors, (b) direction of chain elongation, (c) conservation of the template, and (d) need for a primer.
Question 4.28
Different strands. Explain the difference between the coding strand and the template strand in DNA.
Question 4.29
Family resemblance. Differentiate among mRNA, rRNA and tRNA.
Question 4.30
A code you can live by. What are the key characteristics of the genetic code?
Question 4.31
Encoded sequences.
Write the sequence of the mRNA molecule synthesized from a DNA template strand having the following sequence:
5′–ATCGTACCGTTA–
3′ What amino acid sequence is encoded by the following base sequence of an mRNA molecule? Assume that the reading frame starts at the 5′ end.
5′–UUGCCUAGUGAUUGGAUG–
3′ What is the sequence of the polypeptide formed on addition of poly(UUAC) to a cell-
free protein- synthesizing system?
Question 4.32
A tougher chain. RNA is readily hydrolyzed by alkali, whereas DNA is not. Why?
Question 4.33
A picture is worth a thousand words. Write a reaction sequence showing why RNA is more susceptible to nucleophilic attack than DNA.
Question 4.34
Flowing information. What is meant by the phrase gene expression?
Question 4.35
We can all agree on that. What is a consensus sequence?
Question 4.36
A potent blocker. Cordycepin (3′-deoxyadenosine) is an adenosine analog. When converted into cordycepin 5′-triphosphate, it inhibits RNA synthesis. How does cordycepin 5′-triphosphate block the synthesis of RNA?
Question 4.37
Silent RNA. The code word GGG cannot be deciphered in the same way as can UUU, CCC, and AAA, because poly(G) does not act as a template. Poly(G) forms a triple-
Question 4.38
Sometimes it is not so bad. What is meant by the degeneracy of the genetic code?
Question 4.39
In fact, it can be good. What is the biological benefit of a degenerate genetic code?
Question 4.40
To bring together as associates. Match the components in the right-
|
(a) Replication _____ |
1. RNA polymerase |
|
(b) Transcription _____ |
2. DNA polymerase |
|
(c) Translation _____ |
3. Ribosome |
|
|
4. dNTP |
|
|
5. tRNA |
|
|
6. NTP |
|
|
7. mRNA |
|
|
8. primer |
|
|
9. rRNA |
|
|
10. promoter |
Question 4.41
A lively contest. Match the components in the left-
|
(a) fMet _____ |
1. continuous message |
|
(b) Shine– |
2. removed during processing |
|
(c) intron _____ |
3. the first of many amino acids |
|
(d) exon _____ |
4. joins exons |
|
(e) pre- |
5. joined to make the final message |
|
(f) mRNA _____ |
6. locates the start site |
|
(g) spliceosome _____ |
7. discontinuous message |
Question 4.42
Two from one. Synthetic RNA molecules of defined sequence were instrumental in deciphering the genetic code. Their synthesis first required the synthesis of DNA molecules to serve as templates. Har Gobind Khorana synthesized, by organic-
Question 4.43
Triple entendre. The RNA transcript of a region of T4 phage DNA contains the sequence 5′-AAAUGAGGA-
Question 4.44
A new translation. A transfer RNA with a UGU anticodon is enzymatically conjugated to 14C-
134
5′–UUUUGCCAUGUUUGUGCU–
What is the sequence of the corresponding radiolabeled peptide?
Question 4.45
A tricky exchange. Define exon shuffling and explain why its occurrence might be an evolutionary advantage.
Question 4.46
From one, many. Explain why alternative splicing increases the coding capacity of the genome.
Question 4.47
The unity of life. What is the significance of the fact that human mRNA can be accurately translated in E. coli?
Chapter Integration Problems
Question 4.48
Back to the bench. A protein chemist told a molecular geneticist that he had found a new mutant hemoglobin in which aspartate replaced lysine. The molecular geneticist expressed surprise and sent his friend scurrying back to the laboratory. (a) Why did the molecular geneticist doubt the reported amino acid substitution? (b) Which amino acid substitutions would have been more palatable to the molecular geneticist?
Question 4.49
Eons apart. The amino acid sequences of a yeast protein and a human protein having the same function are found to be 60% identical. However, the corresponding DNA sequences are only 45% identical. Account for this differing degree of identity.
Data Interpretation Problems
Question 4.50
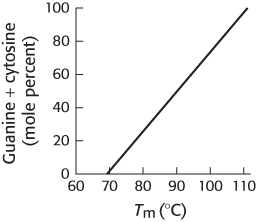
3 is greater than 2. The adjoining illustration graphs the relation between the percentage of GC base pairs in DNA and the melting temperature. Suggest a plausible explanation for these results.
Question 4.51
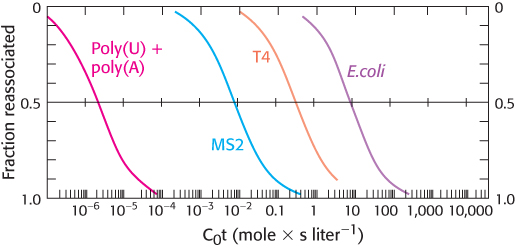
Blast from the past. The illustration below is a graph called a C0t curve (pronounced “cot”). The y-axis shows the percentage of DNA that is double stranded. The x-axis is the product of the concentration of DNA and the time required for the double-
Question 4.52
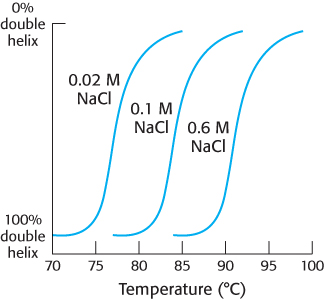
Salt to taste. The graph below shows the effect of salt concentration on melting temperature of bacterial DNA. How does salt concentration affect the melting temperature of DNA? Account for this effect.