4.1 A Nucleic Acid Consists of Four Kinds of Bases Linked to a Sugar–Phosphate Backbone
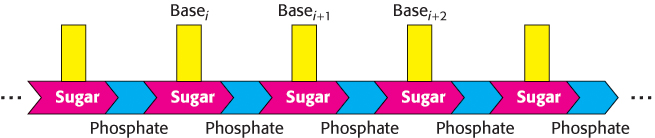
The nucleic acids DNA and RNA are well suited to function as the carriers of genetic information by virtue of their covalent structures. These macromolecules are linear polymers built up from similar units connected end to end (Figure 4.1). Each monomer unit within the polymer is a nucleotide. A single nucleotide unit consists of three components: a sugar, a phosphate, and one of four bases. The sequence of bases in the polymer uniquely characterizes a nucleic acid and constitutes a form of linear information—information analogous to the letters that spell a person’s name.
RNA and DNA differ in the sugar component and one of the bases
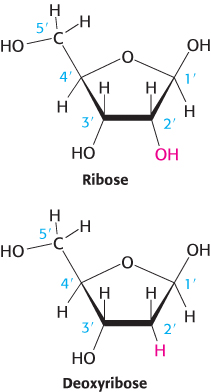
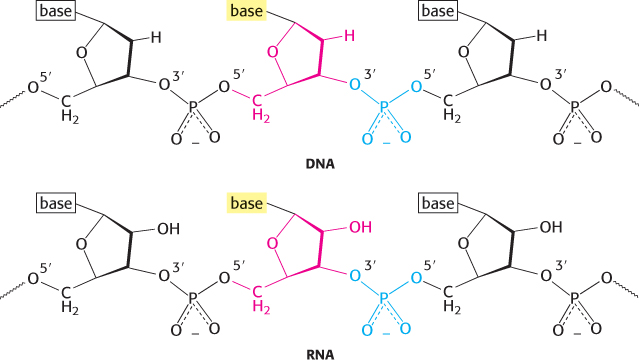
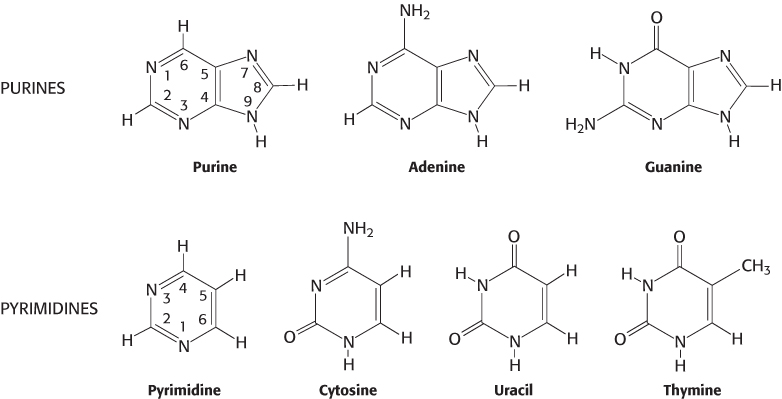
The sugar in deoxyribonucleic acid (DNA) is deoxyribose. The prefix deoxy indicates that the 2′-carbon atom of the sugar lacks the oxygen atom that is linked to the 2′-carbon atom of ribose, as shown in Figure 4.2. Note that sugar carbons are numbered with primes to differentiate them from atoms in the bases. The sugars in both nucleic acids are linked to one another by phosphodiester bridges. Specifically, the 3′-hydroxyl (3′-OH) group of the sugar moiety of one nucleotide is esterified to a phosphate group, which is, in turn, joined to the 5′-hydroxyl group of the adjacent sugar. The chain of sugars linked by phosphodiester bridges is referred to as the backbone of the nucleic acid (Figure 4.3). Whereas the backbone is constant in a nucleic acid, the bases vary from one monomer to the next. Two of the bases of DNA are derivatives of purine—adenine (A) and guanine (G)—and two of pyrimidine—cytosine (C) and thymine (T), as shown in Figure 4.4.
107
Ribonucleic acid (RNA), like DNA, is a long unbranched polymer consisting of nucleotides joined by 3′-to-
Note that each phosphodiester bridge has a negative charge. This negative charge repels nucleophilic species such as hydroxide ions, which are capable of hydrolytic attack on the phosphate backbone. This resistance is crucial for maintaining the integrity of information stored in nucleic acids. The absence of the 2′-hydroxyl group in DNA further increases its resistance to hydrolysis. The greater stability of DNA probably accounts for its use rather than RNA as the hereditary material in all modern cells and in many viruses.
Nucleotides are the monomeric units of nucleic acids
The building blocks of nucleic acids and the precursors of these building blocks play many other roles throughout the cell—
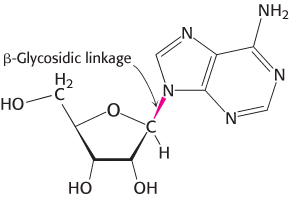
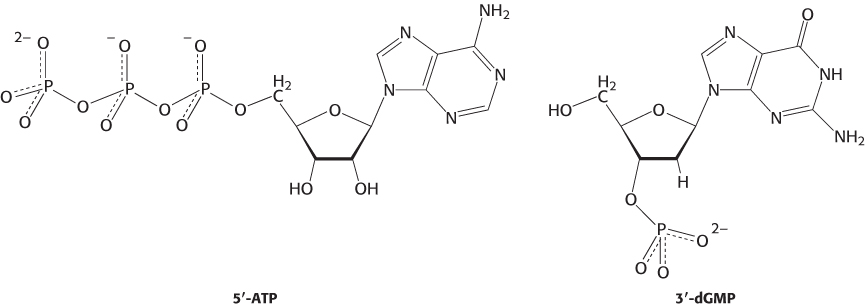
A nucleotide is a nucleoside joined to one or more phosphoryl groups by an ester linkage. Nucleotide triphosphates, nucleosides joined to three phosphoryl groups, are the monomers—
108
This nomenclature does not describe the number of phosphoryl groups or the site of attachment to carbon of the ribose. A more precise nomenclature is also commonly used. A compound formed by the attachment of a phosphoryl group to C-
DNA molecules are very long and have directionality
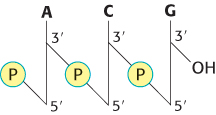
Scientific communication frequently requires the sequence of a nucleic acid—
109
A striking characteristic of naturally occurring DNA molecules is their length. A DNA molecule must comprise many nucleotides to carry the genetic information necessary for even the simplest organisms. For example, the DNA of a virus such as polyoma, which can cause cancer in certain organisms, consists of two paired strands of DNA, each 5100 nucleotides in length. The E. coli genome is a single DNA molecule consisting of two strands of 4.6 million nucleotides each (Figure 4.8).
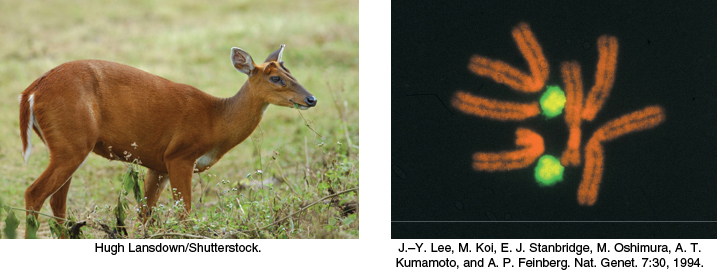
The DNA molecules of higher organisms can be much larger. The human genome comprises approximately 3 billion nucleotides in each strand of DNA, divided among 24 distinct molecules of DNA called chromosomes (22 autosomal chromosomes plus the X and Y sex chromosomes) of different sizes. One of the largest known DNA molecules is found in the Indian muntjac, an Asiatic deer; its genome is nearly as large as the human genome but is distributed on only 3 chromosomes (Figure 4.9). The largest of these chromosomes has two strands of more than 1 billion nucleotides each. If such a DNA molecule could be fully extended, it would stretch more than 1 foot in length. Some plants contain even larger DNA molecules.