Chapter 4
1. A nucleoside is a base attached to a ribose or deoxyribose sugar. A nucleotide is a nucleoside with one or more phosphoryl groups attached to the ribose or deoxyribose.
2. Hydrogen-
3. T is always equal to A, and so these two nucleotides constitute 40% of the bases. G is always equal to C, and so the remaining 60% must be 30% G and 30% C.
4. Nothing, because the base-
5. (a) TTGATC; (b) GTTCGA; (c) ACGCGT; (d) ATGGTA.
6. (a) [T] + [C] = 0.46. (b) [T] = 0.30, [C] = 0.24, and [A] + [G] = 0.46.
7. Stable hydrogen bonding occurs only between GC and AT pairs. Moreover, two purines are too large to fit inside the double helix, and two pyrimidines are too small to form base pairs with each other.
8. The thermal energy causes the chains to wiggle about, which disrupts the hydrogen bonds between base pairs and the stacking forces between bases and thereby causes the strands to separate.
9. The probability that any sequence will appear is 1/4n, where 4 is the number of nucleotides and n is the length of the sequence. The probability of any 15-
10. One end of a nucleic acid polymer ends with a free 5′-hydroxyl group (or a phosphoryl group esterified to the hydroxyl group), and the other end has a free 3′-hydroxyl group. Thus, the ends are different. Two strands of DNA can form a double helix only if the strands are running in different directions—
11. Although the individual bonds are weak, the population of thousands to millions of such bonds provides much stability. There is strength in numbers.
12. There would be too much charge repulsion from the negative charges on the phosphoryl groups. These charges must be countered by the addition of cations.
13. The three forms are the A-
14. 5.88 × 103 base pairs.
15. The diameter of DNA is 20 Å and 1 Å = 0.1 nm, so the diameter is 2 nm. Because 1 μm = 103 nm, the length is 2 × 104 nm. Thus, the axial ratio is 1 × 104.
16. A template is the sequence of DNA or RNA that directs the synthesis of a complementary sequence. A primer is the initial segment of a polymer that is to be extended during elongation.
17. In conservative replication, after 1.0 generation, half of the molecules would be 15N-
18. The nucleotides used for DNA synthesis have the triphosphate attached to the 5′-hydroxyl group with free 3′-hydroxyl groups. Such nucleotides can be utilized only for 5′-to-
19. (a) Tritiated thymine or tritiated thymidine. (b) dATP, dGTP, dCTP, and TTP labeled with 32P in the innermost (α) phosphorus atom.
20. Molecules in parts a and b would not lead to DNA synthesis, because they lack a 3′-OH group (a primer). The molecule in part d has a free 3′-OH group at one end of each strand but no template strand beyond. Only the molecule in part c would lead to DNA synthesis.
21. A retrovirus is a virus that has RNA as its genetic material. However, for the information to be expressed, it must first be converted into DNA, a reaction catalyzed by the enzyme reverse transcriptase. Thus, at least initially, information flow is opposite that of a normal cell: RNA → DNA rather than DNA → RNA.
22. A thymidylate oligonucleotide should be used as the primer. The poly(A) template specifies the incorporation of T; hence, radioactive thymidine triphosphate (labeled in the α phosphoryl group) should be used in the assay.
23. The ribonuclease serves to degrade the RNA strand, a necessary step in forming duplex DNA from the RNA–
24. Treat one aliquot of the sample with ribonuclease and another with deoxyribonuclease. Test these nuclease-
25. Deamination changes the original G · C base pair into a G · U pair. After one round of replication, one daughter duplex will contain a G · C pair and the other duplex will contain an A · U pair. After two rounds of replication, there will be two G · C pairs, one A · U pair, and one A · T pair.
26. (a) 48 = 65,536. In computer terminology, there are 64K 8-
(b) A bit specifies two bases (say, A and C) and a second bit specifies the other two (G and T). Hence, two bits are needed to specify a single nucleotide (base pair) in DNA. For example, 00, 01, 10, and 11 could encode A, C, G, and T. An 8-
(c) A standard CD can hold about 700 megabytes, which is equal to 5.6 × 109 bits. A large number of 8-
A5
27. (a) Deoxyribonucleoside triphosphates versus ribonucleoside triphosphates.
(b) 5′ → 3′ for both.
(c) Semiconserved for DNA polymerase I; conserved for RNA polymerase.
(d) DNA polymerase I needs a primer, whereas RNA polymerase does not.
28. The template strand has a sequence complementary to that of the RNA transcript. The coding strand has the same sequence as that of the RNA transcript except for thymine (T) in place of uracil (U).
29. Messenger RNA encodes the information that, on translation, yields a protein. Ribosomal RNA is the catalytic component of ribosomes, the molecular complexes that synthesize proteins. Transfer RNA is an adaptor molecule, capable of binding a specific amino acid and recognizing a corresponding codon. Transfer RNAs with attached amino acids are substrates for the ribosome.
30. Three nucleotides encode an amino acid; the code is nonoverlapping; the code has no punctuation; the code exhibits directionality; the code is degenerate.
31. (a) 5′-UAACGGUACGAU-
(b) Leu-
(c) Poly(Leu-
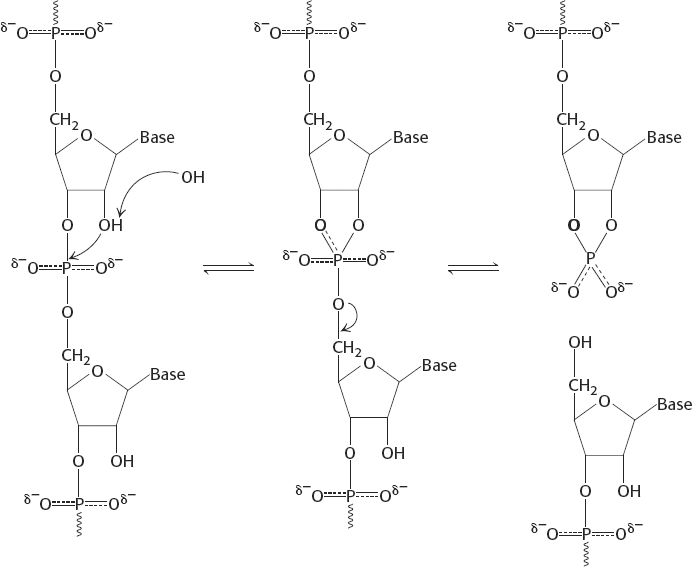
32. The 2′-OH group in RNA acts as an intramolecular nucleophile. In the alkaline hydrolysis of RNA, it forms a 2′-3′ cyclic intermediate.
33.
34. Gene expression is the process of expressing the information of a gene in its functional molecular form. For many genes, the functional information is a protein molecule. Thus, gene expression includes transcription and translation.
35. A nucleotide sequence whose bases represent the most-
36. Cordycepin terminates RNA synthesis. An RNA chain containing cordycepin lacks a 3′-OH group.
37. Only single-
38. Degeneracy of the code refers to the fact that most amino acids are encoded by more than one codon.
39. If only 20 of the 64 possible codons encoded amino acids, then a mutation that changed a codon would likely result in a nonsense codon, leading to termination of protein synthesis. With degeneracy, a nucleotide change might yield a synonym or a codon for an amino acid with similar chemical properties.
40. (a) 2, 4, 8; (b) 1, 6, 10; (c) 3, 5, 7, 9.
41. (a) 3; (b) 6; (c) 2; (d) 5; (e) 7; (f ) 1; (g) 4.
42. Incubation with RNA polymerase and only UTP, ATP, and CTP led to the synthesis of only poly(UAC). Only poly(GUA) was formed when GTP was used in place of CTP.
43. A peptide terminating with Lys (UGA is a stop codon), another containing -Asn-
44. Phe-
45. Exon shuffling is a molecular process that can lead to the generation of new proteins by the rearrangement of exons within genes. Because many exons encode functional protein domains, exon shuffling is a rapid and efficient means of generating new genes.
46. Alternate splicing allows one gene to code for several different but related proteins.
47. It shows that the genetic code and the biochemical means of interpreting the code are common to even very distantly related life forms. It also testifies to the unity of life: that all life arose from a common ancestor.
48. (a) A codon for lysine cannot be changed to one for aspartate by the mutation of a single nucleotide. (b) Arg, Asn, Gln, Glu, Ile, Met, or Thr.
49. The genetic code is degenerate. Of the 20 amino acids, 18 are specified by more than one codon. Hence, many nucleotide changes (especially in the third base of a codon) do not alter the nature of the encoded amino acid. Mutations leading to an altered amino acid are usually more deleterious than those that do not and hence are subject to more stringent selection.
50. GC base pairs have three hydrogen bonds compared with two for AT base pairs. Thus, the higher content of GC means more hydrogen bonds and greater helix stability.
51. C0t value essentially corresponds to the complexity of the DNA sequence—
52. Increasing salt increases the melting temperature. Because the DNA backbone is negatively charged, there is a tendency for charge repulsion to destabilize the helix and cause it to melt. The addition of salt neutralizes the charge repulsion, thereby stabilizing the double helix. The results show that within the parameters of the experiment, more salt results in more stabilization, which is reflected as a higher melting temperature.
A6