PROBLEMS
Question 30.1
Babel fish. Why is protein synthesis also called translation?
Question 30.2
Careful, but not too careful. Why is it crucial that protein synthesis has an error frequency of 10−4?
Question 30.3
Commonalities. What features are common to all tRNA molecules?
Question 30.4
The ol’ two step. What two reaction steps are required for the formation of an aminoacyl-
Question 30.5
The same but different. Why must tRNA molecules have both unique structural features and common structural features?
Question 30.6
Charge it. In the context of protein synthesis, what is meant by an activated amino acid?
Question 30.7
Synthetase mechanism. The formation of isoleucyl-
ATP and 32PPi
tRNA, ATP, and 32PPi
Isoleucine, ATP, and 32PPi
Question 30.8
1 = 2, for sufficiently large values of 1. The energetic equivalent of two molecules of ATP is used to activate an amino acid, yet only one molecule of ATP is used. Explain.
Question 30.9
Sieves. Using threonyl-
Question 30.10
Use all available information. Suggest a reason why there are two classes of aminoacyl-
Question 30.11
Going wobbly. Explain how it is possible that some tRNA molecules recognize more than one codon.
Question 30.12
Light and heavy ribosomes. Ribosomes were isolated from bacteria grown in a “heavy” medium (13C and 15N) and from bacteria grown in a “light” medium (12C and 14N). These 60S ribosomes were added to an in vitro system engaged in protein synthesis. An aliquot removed several hours later was analyzed by density-
Question 30.13
The price of protein synthesis. What is the smallest number of molecules of ATP and GTP consumed in the synthesis of a 200-
Question 30.14
Correct phasing. What is meant by the phrase reading frame?
Question 30.15
Suppressing frameshifts. The insertion of a base in a coding sequence leads to a shift in the reading frame, which in most cases produces a nonfunctional protein. Propose a mutation in a tRNA that might suppress frameshifting.
922
Question 30.16
Tagging a ribosomal site. Design an affinity-
Question 30.17
Viral mutation. An mRNA transcript of a T7 phage gene contains the base sequence
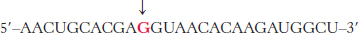
Predict the effect of a mutation that changes the red G to A.
Question 30.18
A new translation. A transfer RNA with a UGU anticodon is enzymatically conjugated to 14C-
5′–UUUUGCCAUGUUUGUGCU–
What is the sequence of the corresponding radiolabeled peptide?
Question 30.19
Two synthetic modes. Compare and contrast protein synthesis by ribosomes with protein synthesis by the solid-
Question 30.20
Triggered GTP hydrolysis. Ribosomes markedly accelerate the hydrolysis of GTP bound to the complex of EF-
Question 30.21
Blocking translation. Devise an experimental strategy for switching off the expression of a specific mRNA without changing the gene encoding the protein or the gene’s control elements.
Question 30.22
Directional problem. Suppose that you have a protein-
At t = 1 minute, you isolate intact protein A from the system, cleave it with trypsin, and isolate the five peptides. Which peptide is most heavily labeled?
At t = 3 minutes, what will be the order of the labeling of peptides from greatest to least?
What does this experiment tell you about the direction of protein synthesis?
Question 30.23
Translator. Aminoacyl-
Question 30.24
A timing device. EF-
Question 30.25
Not just RNA. What are the roles of the protein factors required for protein synthesis?
Question 30.26
Membrane transport. What four components are required for the translocation of proteins across the endoplasmic reticulum membrane?
Question 30.27
Push. Don’t pull. What is the energy source that powers the cotranslational movement of proteins across the endoplasmic reticulum?
Question 30.28
You have to know where to look. Bacterial messenger RNAs usually contain many AUG codons. How does the ribosome identify the AUG specifying initiation?
Question 30.29
Fundamentally the same, yet … List the differences between bacterial and eukaryotic protein synthesis.
Question 30.30
Like a border collie. What is the role of the signal-
Question 30.31
An assembly line. Why is the fact that protein synthesis takes place on polysomes advantageous?
Question 30.32
Match ‘em
|
(a) Initiation ______ |
1. GTP |
|
(b) Elongation ______ |
2. AUG |
|
(c) Termination ______ |
3. fMet |
|
|
4. RRF |
|
|
5.IF2 |
|
|
6. Shine– |
|
|
7. EF- |
|
|
8. Peptidyl transferase |
|
|
9. UGA |
|
|
10. Transformylase |
Question 30.33
Wasted effort? Transfer RNA molecules are quite large, given that the anticodon consists of only three nucleotides. What is the purpose of the rest of the tRNA molecule?
Mechanism Problems
Question 30.34
Evolutionary amino acid choice. Ornithine is structurally similar to lysine except ornithine’s side chain is one methylene group shorter than that of lysine. Attempts to chemically synthesize and isolate ornithinyl-
923
Chapter Integration Problems
Question 30.35
Contrasting modes of elongation. The two basic mechanisms for the elongation of biomolecules are represented in the adjoining illustration. In type 1, the activating group (X) is released from the growing chain. In type 2, the activating group is released from the incoming unit as it is added to the growing chain. Indicate whether each of the following biosyntheses is by means of a type 1 or a type 2 mechanism:
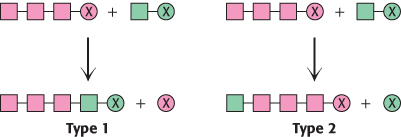
Glycogen synthesis
Fatty acid synthesis
C5 → C10 → C15 in cholesterol synthesis
DNA synthesis
RNA synthesis
Protein synthesis
Question 30.36
Enhancing fidelity. Compare the accuracy of DNA replication, RNA synthesis, and protein synthesis. Which mechanisms are used to ensure the fidelity of each of these processes?
Question 30.37
Déjà vu. Which protein in G-
Question 30.38
Family resemblance. Eukaryotic elongation factor 2 is inhibited by ADP ribosylation catalyzed by diphtheria toxin. What other G proteins are sensitive to this mode of inhibition?
Question 30.39
The exceptional E. coli. In contrast with E. coli, most bacteria do not have a full complement of aminoacyl-
Question 30.40
The final step. What aspect of primary structure allows the transfer of linear nucleic acid information into the functional three-
Data Interpretation Problems
Question 30.41
Helicase helper. The initiation factor eIF-
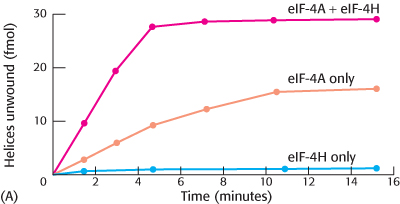
What are the effects on eIF-
4 helicase activity in the presence of eIF- 4H? Why did measuring the helicase activity of eIF-
4H alone serve as an important control? 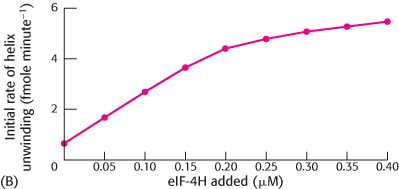
The initial rate of helicase activity of 0.2 μM of eIF-
4 was then measured with varying amounts of eIF- 4H (graph B). What ratio of eIF- 4H to eIF- 4 yielded optimal activity? 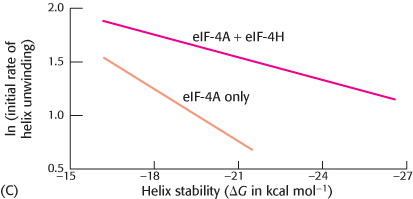 [Data from N. J. Richter, G. W. Rodgers, Jr., J. O. Hensold, and W. C. Merrick. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H., J. Biol. Chem. 274:35415–
[Data from N. J. Richter, G. W. Rodgers, Jr., J. O. Hensold, and W. C. Merrick. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H., J. Biol. Chem. 274:35415–35424, 1999.] Next, the effect of RNA–
RNA helix stability on the initial rate of unwinding in the presence and absence of eIF- 4H was tested (graph C). How does the effect of eIF- 4H vary with helix stability? How might eIF-
4H affect the helicase activity of eIF- 4A?
924
Question 30.42
Size separation. The protein-
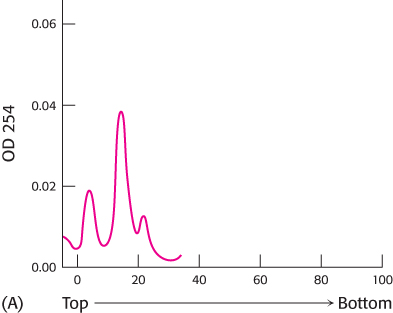
What do the three peaks of absorbance in graph A represent?
The experiment was repeated except that, this time, the RNase treatment was omitted.
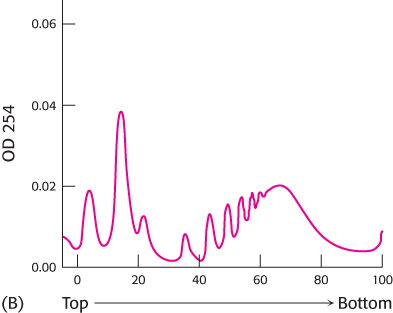
Why is the centrifugation pattern in graph B more complex? What do the series of peaks near the bottom of the centrifuge tube represent?
Before the isolation of the protein-
synthesizing machinery, the cells were grown in low concentrations of oxygen (hypoxic conditions). Again the experiment was repeated without RNase treatment (graph C). 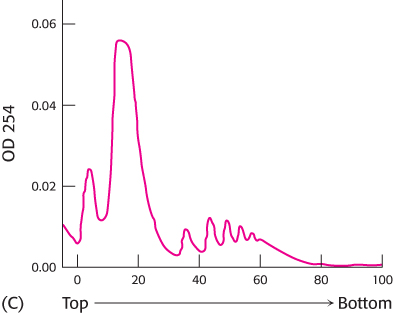 [Data from M. Koritzinsky et al., EMBO J. 25:1114–
[Data from M. Koritzinsky et al., EMBO J. 25:1114–1125, 2006.] What is the effect of growing cells under hypoxic conditions?