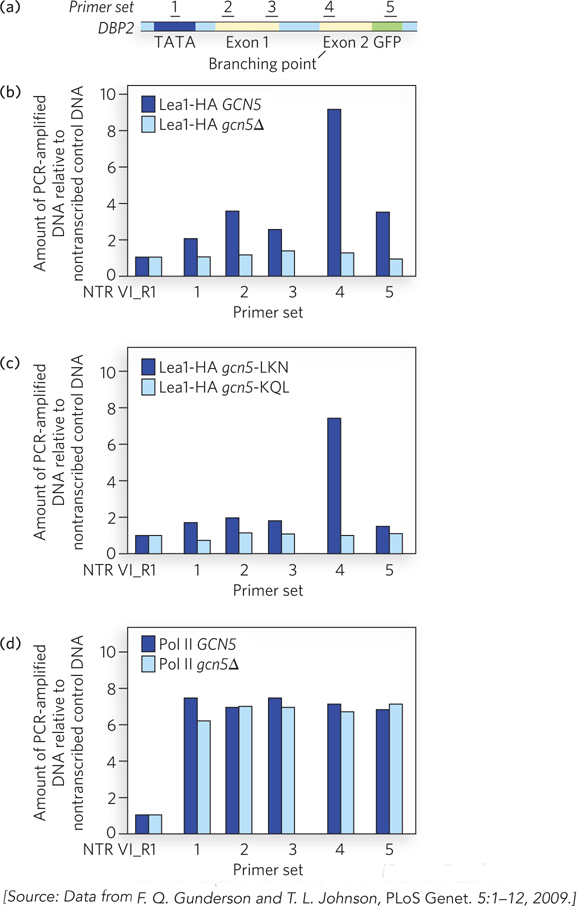
FIGURE 1 Gcn5 activity helps recruit spliceosomal components to DBP2 pre- 1- i- 1- d-