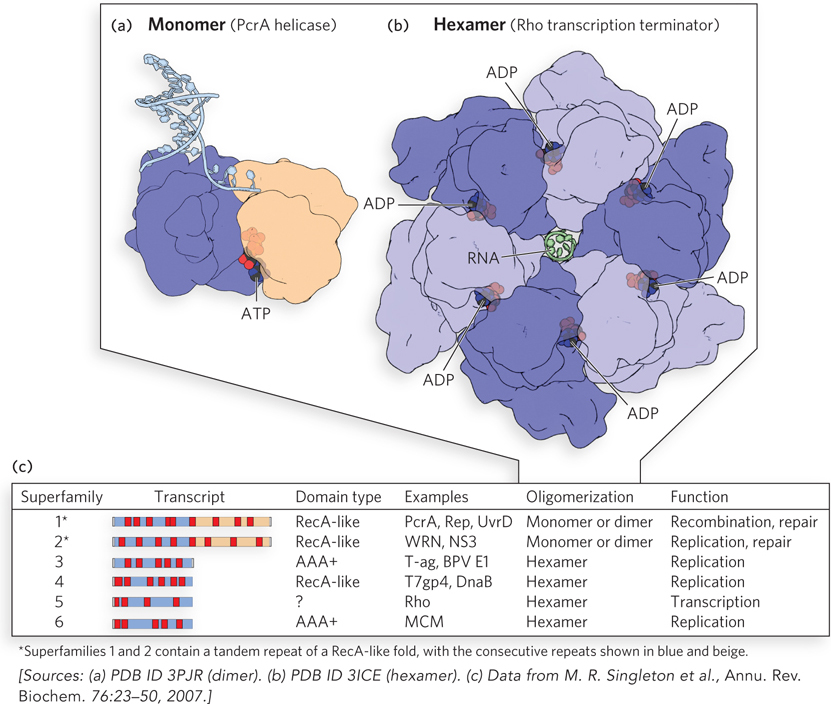
DNA helicases. (a) Enzymes of helicase superfamilies 1 and 2 (SF1 and SF2) have a conserved core structure, the RecA- e- t- l-