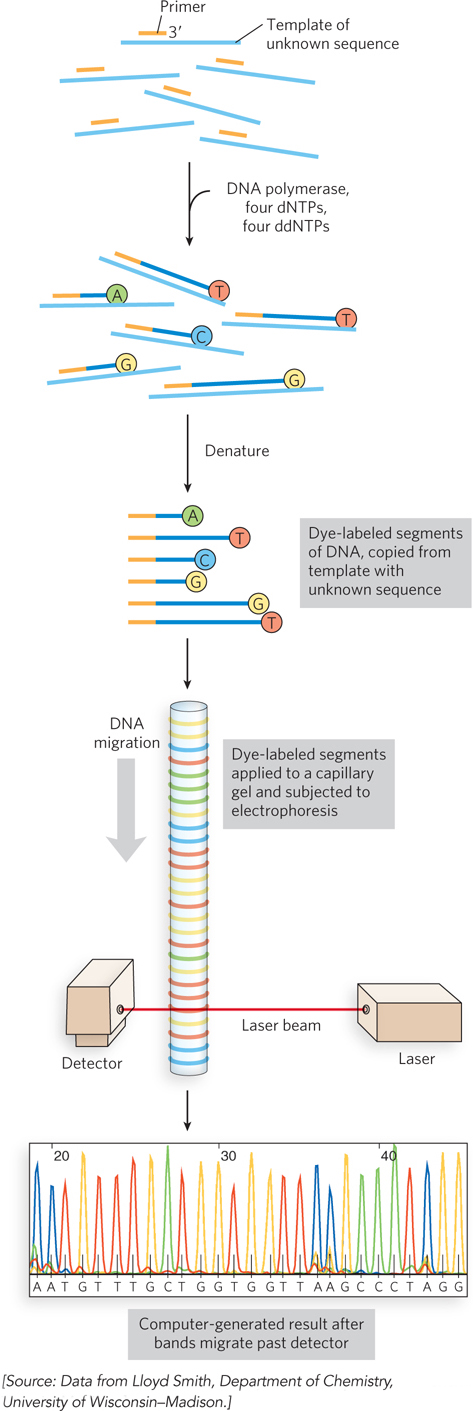
Automation of DNA sequencing reactions. In the Sanger method, each ddNTP can be linked to a fluorescent (dye) molecule that gives the same color to all the fragments terminating in that nucleotide, a different color for each nucleotide. All four labeled ddNTPs are added together. The resulting colored DNA fragments are separated by size in an electrophoretic gel in a capillary tube (a refinement of gel electrophoresis that allows for faster separations). All fragments of a given length migrate through the capillary gel together in a single band, and the color associated with each band is detected with a laser beam. The DNA sequence is read by identifying the color sequences in the bands as they pass the detector. This information is fed directly to a computer, and the sequence is determined. The amount of fluorescence in each band is represented as a peak in the computer output. Here, the nucleotide colors reflect the dyes actually used in the method, and thus deviate from the standard nucleotide colors used in other figures.