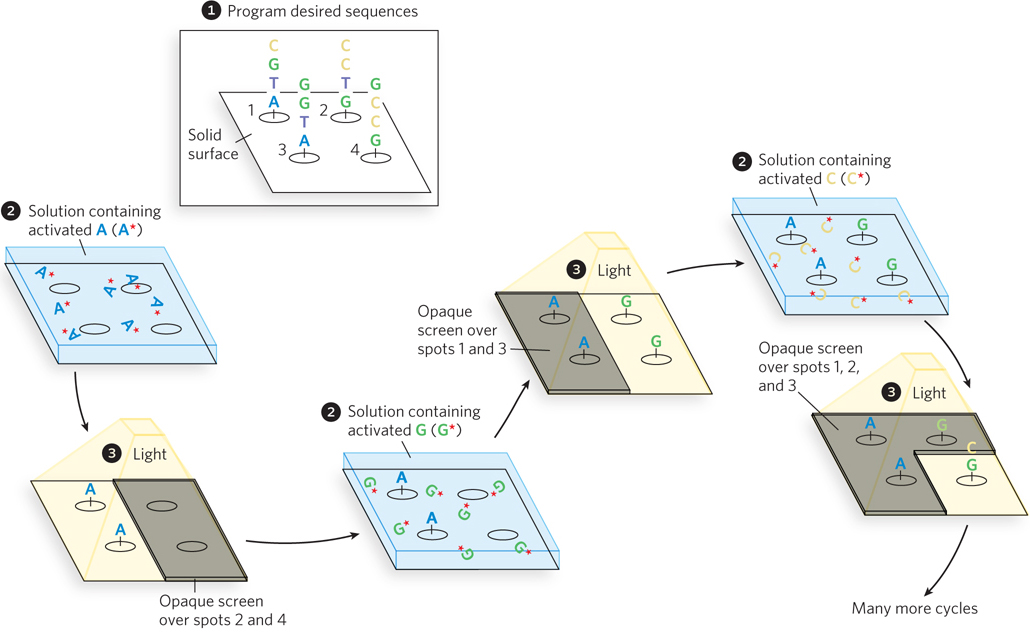
Photolithography to create a DNA microarray.  A computer is programmed with the desired oligonucleotide sequences.
A computer is programmed with the desired oligonucleotide sequences.  The reactive groups, attached to a solid surface, are initially rendered inactive by photoactive blocking groups, which can be removed by a flash of light. An opaque screen blocks the light from certain groups, preventing their activation. Other areas or “spots” are exposed.
The reactive groups, attached to a solid surface, are initially rendered inactive by photoactive blocking groups, which can be removed by a flash of light. An opaque screen blocks the light from certain groups, preventing their activation. Other areas or “spots” are exposed.  A solution containing one activated nucleotide (e.g., A*) is washed over the spots. The 5′ hydroxyl of the nucleotide is blocked to prevent unwanted reactions, and the nucleotide links to the surface groups at the appropriate spots through its 3′ hydroxyl. The surface is washed successively with solutions containing each remaining activated nucleotide (G*, C*, T*). The 5′-blocking groups on each nucleotide limit the reactions to addition of one nucleotide at a time, and these groups can also be removed by light. Once each spot has one nucleotide, a second nucleotide can be added to extend the nascent oligonucleotide at each spot, using screens and light to ensure that the correct nucleotides are added at each spot in the correct sequence. This continues until the required sequences are built up on each spot on the surface.
A solution containing one activated nucleotide (e.g., A*) is washed over the spots. The 5′ hydroxyl of the nucleotide is blocked to prevent unwanted reactions, and the nucleotide links to the surface groups at the appropriate spots through its 3′ hydroxyl. The surface is washed successively with solutions containing each remaining activated nucleotide (G*, C*, T*). The 5′-blocking groups on each nucleotide limit the reactions to addition of one nucleotide at a time, and these groups can also be removed by light. Once each spot has one nucleotide, a second nucleotide can be added to extend the nascent oligonucleotide at each spot, using screens and light to ensure that the correct nucleotides are added at each spot in the correct sequence. This continues until the required sequences are built up on each spot on the surface.
 A computer is programmed with the desired oligonucleotide sequences.
A computer is programmed with the desired oligonucleotide sequences.  The reactive groups, attached to a solid surface, are initially rendered inactive by photoactive blocking groups, which can be removed by a flash of light. An opaque screen blocks the light from certain groups, preventing their activation. Other areas or “spots” are exposed.
The reactive groups, attached to a solid surface, are initially rendered inactive by photoactive blocking groups, which can be removed by a flash of light. An opaque screen blocks the light from certain groups, preventing their activation. Other areas or “spots” are exposed.  A solution containing one activated nucleotide (e.g., A*) is washed over the spots. The 5′ hydroxyl of the nucleotide is blocked to prevent unwanted reactions, and the nucleotide links to the surface groups at the appropriate spots through its 3′ hydroxyl. The surface is washed successively with solutions containing each remaining activated nucleotide (G*, C*, T*). The 5′-blocking groups on each nucleotide limit the reactions to addition of one nucleotide at a time, and these groups can also be removed by light. Once each spot has one nucleotide, a second nucleotide can be added to extend the nascent oligonucleotide at each spot, using screens and light to ensure that the correct nucleotides are added at each spot in the correct sequence. This continues until the required sequences are built up on each spot on the surface.
A solution containing one activated nucleotide (e.g., A*) is washed over the spots. The 5′ hydroxyl of the nucleotide is blocked to prevent unwanted reactions, and the nucleotide links to the surface groups at the appropriate spots through its 3′ hydroxyl. The surface is washed successively with solutions containing each remaining activated nucleotide (G*, C*, T*). The 5′-blocking groups on each nucleotide limit the reactions to addition of one nucleotide at a time, and these groups can also be removed by light. Once each spot has one nucleotide, a second nucleotide can be added to extend the nascent oligonucleotide at each spot, using screens and light to ensure that the correct nucleotides are added at each spot in the correct sequence. This continues until the required sequences are built up on each spot on the surface.