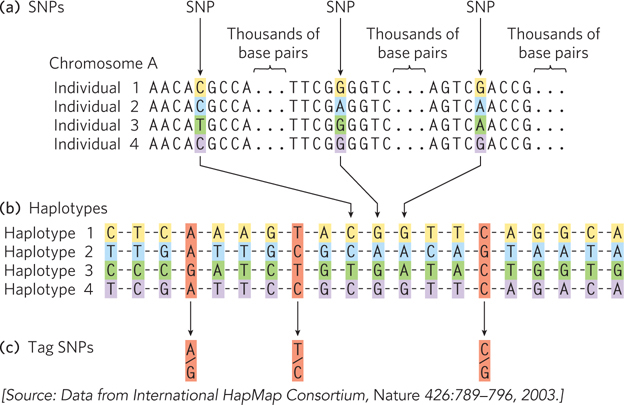
Haplotype identification. (a) Positions in the human genome where single nucleotide polymorphisms occur are often identified in genomic samples. The SNPs can be in any part of the genome, whether or not part of a known gene. (b) Groups of these SNPs are compiled into a haplotype. The SNPs will vary in the overall human population, such as in the four fictional individuals shown here, but the SNPs chosen to define a haplotype are often the same in most individuals of a particular population. (c) A few single nucleotide polymorphisms are chosen as haplotype- A- T- G- T-