PROBLEMS
Question 5.1
Protein A has a binding site for ligand X with a Kd of 106 m. Protein B has a binding site for ligand X with a Kd of 109 m. Which protein has a higher affinity for ligand X? Explain your reasoning. Convert Kd to Ka for both proteins.
Question 5.2
The lactose (Lac) repressor has a binding site for DNA and binds to a specific DNA site with a Kd of approximately 10−10 m. The Lac repressor also has a binding site for the galactoside allolactose. When the Lac repressor interacts with allolactose, it dissociates from the DNA. When allolactose is bound to the Lac repressor, does the Kd for the repressor’s specific DNA binding site increase or decrease? Explain.
Question 5.3
The Lac repressor interacts with DNA through its helix-
Question 5.4
The binding of any protein to DNA invariably involves the displacement of other atoms or molecules. What types of atoms or molecules are most commonly displaced?
Question 5.5
When a protein binds to DNA nonspecifically, with which parts of the DNA does the protein generally interact? When a protein binds to DNA at a site defined by a particular nucleotide sequence, with which parts of the DNA does the protein generally interact?
Question 5.6
Which of the following situations would produce negative cooperativity? Explain your reasoning in each case.
The protein has multiple subunits, each with a single ligand-
binding site. The binding of ligand to one site decreases the binding affinity of other sites for the ligand. The protein is a single polypeptide with two independent ligand-
binding sites, each having a different affinity for the ligand. The protein is a single polypeptide with a single ligand-
binding site. As purified, the protein preparation is heterogeneous, containing some protein molecules that are partially denatured and thus have a lower binding affinity for the ligand.
Question 5.7
To approximate the actual concentration of enzymes in a bacterial cell, assume that the cell contains equal concentrations of 1,000 different enzymes in solution in the cytosol and that each protein has a molecular weight of 100,000. Assume also that the bacterial cell is a cylinder (diameter 1.0 μm, height 2.0 μm), that the cytosol (specific gravity 1.20) is 20% soluble protein by weight, and that the soluble protein consists entirely of enzymes. Calculate the average molar concentration of each enzyme in this hypothetical cell.
Question 5.8
Which of the following effects would be brought about by any enzyme catalyzing the simple reaction 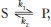 where Keq = [P]/[S]? (a) Decreased Keq; (b) increased k1; (c) increased Keq; (d) increased ΔG‡; (e) decreased ΔG‡; (f) more negative ΔG′°; (g) increased k2.
where Keq = [P]/[S]? (a) Decreased Keq; (b) increased k1; (c) increased Keq; (d) increased ΔG‡; (e) decreased ΔG‡; (f) more negative ΔG′°; (g) increased k2.
Question 5.9
In the late nineteenth century, the famous chemist Emil Fischer proposed that an enzyme should possess a pocket that is complementary in shape to the substrate it interacts with in its catalytic function. This “lock and key” hypothesis was highly influential at the time. Although Fischer made many important contributions to the development of enzymology, this particular idea was largely wrong. Explain why.
Question 5.10
If an irreversible inhibitor inactivates an enzyme, what is the effect on that enzyme’s kcat and Km?
Question 5.11
At what substrate concentration would an enzyme with a kcat of 30.0 s−1 and a Km of 0.0050 m operate at one-
quarter of its maximum rate? Determine the fraction of Vmax that would be achieved at the following substrate concentrations: [S] = ½Km, 2Km, and 10Km.
An enzyme that catalyzes the reaction X ⇌ Y is isolated from two bacterial species. The two enzymes have the same Vmax but different Km values for the substrate X. Enzyme A has a Km of 2.0 μm, and enzyme B has a Km of 0.5 μm. The plot below shows the kinetics of reactions carried out with the same concentration of each enzyme and with [X] = 1 μm. Which curve corresponds to which enzyme?
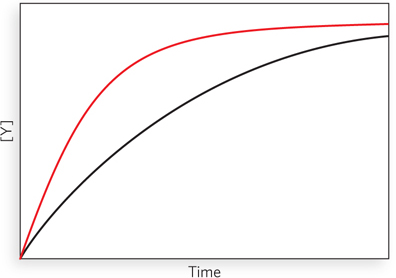
170
Question 5.12
A research group discovers a new enzyme, which they call happyase, that catalyzes the chemical reaction HAPPY ⇌ SAD. The researchers begin to characterize the enzyme.
In the first experiment, with a total concentration of enzyme, [Et], at 4 nM, they find that Vmax = 1.6 μm s−1. Based on this experiment, what is the kcat for happyase? (Include the appropriate units.)
In another experiment, with [Et] at 1 nM and [HAPPY] at 30 μm, the researchers find that V0 = 300 nM s−1. What is the measured Km of happyase for its substrate HAPPY? (Include the appropriate units.)
Further research shows that the supposedly purified happyase used in the first two experiments was actually contaminated with a reversible inhibitor called ANGER. When ANGER is carefully removed from the happyase preparation, and the two experiments are repeated, the measured Vmax in (a) increases to 4.8 μm s−1, and the measured Km in (b) is 15 μm. For the inhibitor ANGER present in the original preparation, calculate the values of α and α′ (see Highlight 5-1).
Based on the information given above, what type of inhibitor is ANGER?
Question 5.13
An enzyme is discovered that catalyzes the reaction A ⇌ B. Researchers find that the Km for substrate A is 4 μm, and the kcat is 20 min−1.
In an experiment, [A] = 6 mM, and the initial velocity, V0, is measured as 480 nM min−1. What is the [Et] used in the experiment?
In another experiment, [Et] = 0.5 nM, and the measured V0 = 5 nM min−1. What is the [A] used in the experiment?
The compound Z is found to be a very strong competitive inhibitor of the enzyme. In an experiment with the same [Et] as in part (a) but a different [A], an amount of Z is added that produces an α = 10. This reduces V0 to 240 nM min−1. What is the [A] in this experiment?
Question 5.14
Although graphical methods are available for accurate determination of the Vmax and Km of an enzyme-

Question 5.15
The bacterial RuvB protein, a DNA translocase, belongs to helicase superfamily 6. RuvB functions as a circular hexameric complex with a central opening. Duplex DNA is bound in the opening. RuvB is also an ATPase, and it moves along the DNA when ATP is hydrolyzed. An amino acid substitution in the ATP-
Question 5.16
When eukaryotic DNA replication is prematurely halted, due to DNA damage or other causes, two proteins—
Question 5.17
A biochemist is purifying a new DNA helicase encoded by a pathogenic virus. The enzyme activity is readily detected after the first several purification steps that generate fractions a, b, and c, each with successively greater purity. The researcher then puts fraction c on an ion-
171