DATA ANALYSIS PROBLEM
Cohen, S.N., A.C.Y. Chang, H.W. Boyer, and R.B. Helling. 1973. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 70:3240–
Question 7.20
The first recombinant DNA plasmid that combined two different biological activities from different sources and was readily transferred into bacteria was reported in 1973 by the research groups of Stanley Cohen and Herbert Boyer. In this work, the investigators began with the large, naturally occurring circular plasmid R6-
The ultimate objective was to combine pSC101 with fragments from R6-
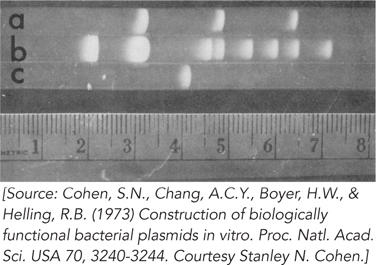
From this pattern, how many EcoRI sites are present in each of the three plasmids? (Note that three small DNA bands from R6-
5 are not visible on this gel.) Why does the brightness of the bands decrease from left to right?
In the R6-
5 lane (lane b), the second largest band seems to break the general pattern in that it is brighter than the band to its left. How might this be explained? The three bands in the pSC102 lane (lane a) comigrate with bands in the R6-
5 lane. Explain.
The plasmids pSC101 and pSC102 were cleaved with EcoRI, and the fragments from both plasmids were mixed together and treated with DNA ligase. The ligated mixture was then used to transform E. coli, and the cells were grown on plates containing both kanamycin and tetracycline. Colonies appeared, and all of them contained a new plasmid, named pSC105 (∼16,000 bp), that conferred resistance to kanamycin and tetracycline. Next, the plasmids pSC101, pSC102, and pSC105 were cleaved with EcoRI and subjected to gel electrophoresis. The results are shown in Figure 2. Electrophoresis proceeded, and fragment sizes decrease, from left to right (lane a is pSC105; lane b, pSC101 + pSC102; lane c, pSC102; lane d, pSC101).
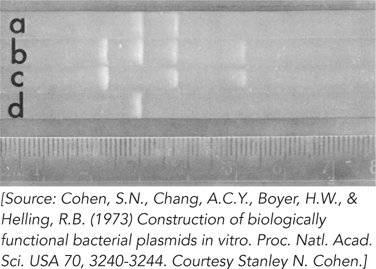
Which of the two fragments of pSC105 (lane a) contains the gene for tetracycline resistance?
Which of the two fragments of pSC105 contains the gene encoding kanamycin resistance?
What is the approximate size of the second (smaller) EcoRI cleavage fragment of pSC105?
How many phosphodiester bonds were created by DNA ligase to produce the circular plasmid pSC105?
The largest and smallest EcoRI fragments of pSC101 (lane d) do not appear in pSC105, although they were present in the ligation mixture that gave rise to it. Why were these DNA fragments not incorporated into the recombinant plasmid?
257