PROBLEMS
Question 8.1
Three different but overlapping BAC clones (see Chapter 7) were digested with the restriction enzyme EcoRI and the fragments separated on an agarose gel, as shown in the figure below. Only the cloned DNA (not the plasmid vector) is shown. Order these three clones into a contig, and label the contig with the location of the EcoRI restriction sites and the distances between them.
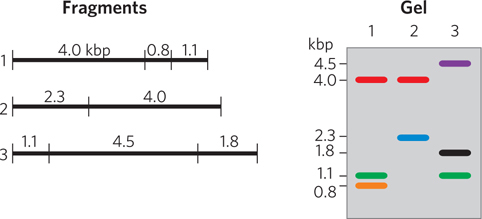
Question 8.2
A researcher compares the amino acid sequences of cytochrome c from four vertebrates: sheep, dog, rabbit, and kangaroo. The amino acid differences among the four species are shown in the difference matrix below. From this information, build a simple evolutionary tree expressing the apparent relationships among these organisms. The branch lengths do not have to represent any measure of time.
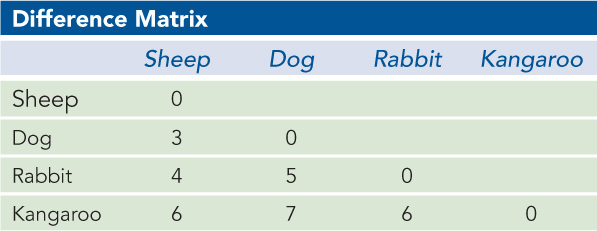
Question 8.3
In random shotgun sequencing, cloned genomic DNA from an organism is sequenced at random. Sequencing requires the use of a primer targeted to a known sequence, which can then be extended to reveal the entire sequence by the traditional Sanger method (see Chapter 7). If the researcher has no sequence information, how can any genome sequences be targeted by primers to initiate the sequencing reactions?
Question 8.4
A hypothetical protein is found in orangutans, chimpanzees, and humans that has the following sequences (red indicates the amino acid residue differences; dashes indicate a deletion—
|
Human: |
ATSAAGYDEWEGGKVLIHL–-KLQNRGALLELDIGAV |
|
Orangutan: |
ATSAAGWDEWEGGKVLIHLDGKLQNRGALLELDIGAV |
|
Chimpanzee: |
ATSAAGWDEWEGGKILIHLDGKLQNRGALLELDIGAV |
What is the most likely sequence of the protein present in the last common ancestor of chimpanzees and humans?
Question 8.5
For defining a cell’s transcriptome, RNA-
How does the sequencing yield information about the levels of specific RNAs in a cell?
Why must rRNA be removed from most samples before conversion of the cellular RNA to cDNA?
Question 8.6
A comparison between two homologous chromosomes in two closely related mammals reveals synteny over most of the length of the chromosomes. However, researchers encounter a segment of about 2,300 bp in mammal Y that is not present in mammal X. What evolutionary processes could account for this difference?
Question 8.7
In large genome sequencing projects, the initial data usually reveal gaps where no sequence information has been obtained. To close the gaps, DNA primers complementary to the 5′-ending strand (i.e., identical to the sequence of the 3′-ending strand) at the end of each contig are especially useful. Explain how these primers might be used.
Question 8.8
In proteomics work, two-
294
Question 8.9
You have just isolated a new protein and you want to know what other proteins it might interact with. Using genetic engineering, you construct a tagged version of your protein, and then use the tag to precipitate it (sometimes called pulling it down) along with other proteins it interacts with in the cell. You subject the precipitated protein mixture to mass spectrometry and identify several hundred different peptides from over a hundred different proteins. Do all of these proteins necessarily interact functionally with your newly isolated protein? What factors might lead noninteracting proteins to precipitate with your protein?
Question 8.10
You are a gene hunter, trying to find the genetic basis for a rare inherited disease. Examination of six pedigrees of families affected by the disease provides inconsistent results. For two of the families, the disease is co-
Question 8.11
In two closely related bacterial species, a cluster of five genes is found on the chromosome, with the genes arranged in the same order. However, in species B, gene X, not present in species A, is found between genes 2 and 3 of the five-
Question 8.12
Native American populations in North and South America have mitochondrial DNA haplotypes that can be traced to populations in northeast Asia. The Aleut and Eskimo populations in the far northern parts of North America possess a subset of the same haplotypes that link other Native Americans to Asia, and they have several additional haplotypes that can also be traced to Asian origins but are not found in native populations in other parts of the Americas. Provide a possible explanation.
Question 8.13
In the evolutionary line leading to modern humans, bottleneck periods occurred in which relatively few individuals survived. All humans living today carry mitochondrial DNA markers derived from a single female, sometimes referred to as mitochondrial Eve. All male humans living today have Y chromosome markers from an ancestor called Y chromosome Adam. Did Y chromosome Adam carry mitochondrial DNA from mitochondrial Eve?
Question 8.14
DNA (haplotypes) originating from Denisovans can be found in the genomes of Australian Aborigines and Melanesian islanders. However, the same DNA markers are not found in the genomes of people native to Africa. Provide an explanation.