DATA ANALYSIS PROBLEM
Ksiazek, T.G., D.V.M. Ksiazek, D. Erdman, C.S. Goldsmith, S.R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J.A. Comer, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–
Question 8.15
T. G. Ksiazek and other members of the SARS Working Group described their discovery of the SARS virus and its identification as a novel coronavirus. They identified the virus as a coronavirus through electron microscopy, and then confirmed it with a sequence analysis of PCR-
The PCR method for amplifying the coronavirus genomic sample requires the use of reverse transcriptase. Why?
To amplify sequences from a new virus with an unsequenced genome, what considerations would go into the design of appropriate PCR primers?
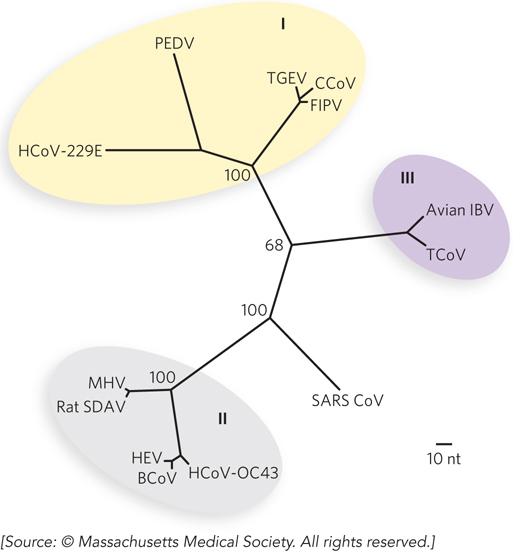
A sequence alignment involving a 405 bp segment of the DNA polymerase gene in four coronaviruses, BCoV, HEV, SARS CoV, and TGEV, is given on the facing page—
the alignment that Ksiazek and colleagues used to generate the phylogenetic tree below (this includes coronaviruses not discussed in this problem; nt = nucleotides). In the alignment, nucleotide polymorphisms relative to the bovine coronavirus sequence (bcov) are shown in red. How many sequence differences exist between the genome segments from HEV (hev) and BCoV? How many between BCoV and SARS CoV (sars)? How many between TGEV (tgev) and SARS CoV? Which two coronaviruses are most closely related? Are your counts in general agreement with the phylogenetic tree?
295
296
| 1 | 60 | ||
|
bcov.pol.seq |
TCGTGCTATGCCAAACATACTACGTATTGTTAGTAGTCTGGTTTTGGCTCGAAAACATGA |
||
|
hev.pol.seq |
TCGTGCTATGCCAAACATACTACGTATTGTTAGTAGTCTGGTATTGGCCCGAAAACATGA |
||
|
sars.pol.seq |
CAGAGCCATGCCTAACATGCTTAGGATAATGGCCTCTCTTGTTCTTGCTCGCAAACATAA |
||
|
tgev.pol.seq |
CCGTGCTTTACCTAATATGATTAGAATGGCTTCTGCCATGATATTAGGTTCTAAGCATGT |
||
| 61 | 120 | ||
|
bcov.pol.seq |
GGCATGTTGTTCGCAAAGCGATAGGTTTTATCGACTTGCGAATGAATGCGCACAAGTTCT |
||
|
hev.pol.seq |
GGCATGTTGTTCGCAAAGCGATAGGTTTTATCGACTTGCGAATGAATGCGCACAAGTTCT |
||
|
sars.pol.seq |
CACTTGCTGTAACTTATCACACCGTTTCTACAGGTTAGCTAACGAGTGTGCGCAAGTATT |
||
|
tgev.pol.seq |
TGGTTGTTGTACACATAATGATAGGTTCTACCGCCTCTCCAATGAGTTAGCTCAAGTACT |
||
| 121 | 180 | ||
|
bcov.pol.seq |
GAGTGAAATTGTTATGTGTGGTGGCTGTTATTATGTTAAGCCTGGTGGCACTAGTAGTGG |
||
|
hev.pol.seq |
TAGTGAAATTGTTATGTGTGGTGGCTGTTATTATGTTAAGCCTGGTGGCACTAGTAGTGG |
||
|
sars.pol.seq |
AAGTGAGATGGTCATGTGTGGCGGCTCACTATATGTTAAACCAGGTGGAACATCATCCGG |
||
|
tgev.pol.seq |
CACAGAAGTTGTGCATTGCACAGGTGGTTTTTATTTTAAACCTGGTGGTACAACTAGCGG |
||
| 181 | 240 | ||
|
bcov.pol.seq |
TGATGCAACTACTGCTTTTGCTAATTCAGTTTTTAACATATGTCAAGCTGTTTCAGCCAA |
||
|
hev.pol.seq |
TGATGCAACTACTGCTTTTGCTAATTCAGTCTTTAACATATGTCAAGCTGTTTCAGCCAA |
||
|
sars.pol.seq |
TGATGCTACAACTGCTTATGCTAATAGTGTCTTTAACATTTGTCAAGCTGTTACAGCCAA |
||
|
tgev.pol.seq |
TGATGGTACTACAGCATATGCTAACTCTGCTTTTAACATCTTTCAAGCTGTTTCTGCTAA |
||
| 241 | 300 | ||
|
bcov.pol.seq |
TGTATGTGCTTTAATGTCATGCAATGGTAATAAGATTGAAGATTTGAGTATACGTGCTCT |
||
|
hev.pol.seq |
TGTATGTTCCTTAATGTCATGCAATGGCAATAAGATTGAAGATTTGAGTATACGTGCTCT |
||
|
sars.pol.seq |
TGTAAATGCACTTCTTTCAACTGATGGTAATAAGATAGCTGACAAGTATGTCCGCAATCT |
||
|
tgev.pol.seq |
TGTTAATAAGCTTTTGGGGGTTGATTCAAACGCTTGTAACAACGTTACAGTAAAATCCAT |
||
| 301 | 360 | ||
|
bcov.pol.seq |
TCAGAAGCGCTTATACTCACATGTGTATAGAAGTGATATGGTTGATTCAACCTTTGTCAC |
||
|
hev.pol.seq |
TCAGAAGCGTTTATACTCACATGTGTATAGAAGTGATATGGTTGATTCAACCTTTGTCAC |
||
|
sars.pol.seq |
ACAACACAGGCTCTATGAGTGTCTCTATAGAAATAGGGATGTTGATCATGAATTCGTGGA |
||
|
tgev.pol.seq |
ACAACGTAAAATTTACGATAATTGTTATCGTAGTAGCAGCATTGATGAAGAATTTGTTGT |
||
| 361 | 406 | ||
|
bcov.pol.seq |
AGAATATTATGAATTTTTAAATAAGCATTTTAGTATGATGATTTTG |
||
|
hev.pol.seq |
AGAATATTATGAATTTTTAAATAAGCATTTTAGTATGATGATTTTG |
||
|
sars.pol.seq |
TGAGTTTTACGCTTACCTGCGTAAACATTTCTCCATGATGATTCTT |
||
|
tgev.pol.seq |
TGAGTACTTTAGTTATTTGAGAAAACACTTTTCTATGATGATTTTA |
||
|
[Source: Sequence alignment courtesy of Dr. Ann Palmenberg, Department of Biochemistry and the Institute for Molecular Virology, University of Wisconsin–Madison.] |
|||