Chapter 8
Question 8.1
Question 8.2
S-
Question 8.3
The genomic DNA fragments are cloned into plasmid vectors. Although the sequence of the cloned DNA is not known, the plasmid sequences immediately adjacent to a DNA fragment are known. A single primer is used, targeted to the plasmid sequence near one end of the cloning site, and sequencing of each clone is initiated from that point.
Question 8.4
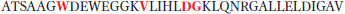
Question 8.5
(a) Modern sequencing of the cDNAs generated in RNA-
Question 8.6
The possibilities include gene duplication, horizontal gene transfer, and transposon insertion.
Question 8.7
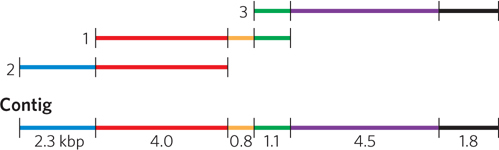
The primers can be used to probe libraries containing long genomic clones to identify contig ends that lie close to each other. If the contigs are close enough, the primers can be used in PCR to directly amplify the intervening DNA separating the contigs, which can then be cloned and sequenced.
Question 8.8
If the same procedure were used in both dimensions, all the proteins would form a single diagonal line in the final gel, and much of the potential for separation would be wasted. Using different protein properties in the two electrophoresis steps effectively spreads the proteins over the entire gel.
Question 8.9
Proteins may precipitate nonspecifically for many reasons. In many cases, the precipitation is simply a property of the proteins and the conditions used. The interaction might also be indirect, with both the target protein and the nonspecific precipitating protein interacting in common with some other macromolecule, such as another protein or nucleic acid. Researchers focus on the precipitating proteins that are most abundant and confirm the putative interaction with additional experiments.
Question 8.10
The same disease condition can be caused by defects in two or more genes, which are on different chromosomes.
Question 8.11
If gene X has no relationship to any other gene in species B, it may have arisen by horizontal gene transfer. If gene X is homologous to gene 2, it may have arisen by gene duplication.
Question 8.12
The pattern of haplotypes in the Aleut and Eskimo populations suggests that their ancestors migrated into the American Arctic regions in a separate migration from the one that led to the populating of the rest of North and South America.
Question 8.13
Yes. Mitochondrial Eve lived thousands of years before Y chromosome Adam. Given that all modern humans (male and female) inherited their mitochondrial DNA from Eve, Adam must have had that DNA as well.
Question 8.14
Interbreeding between Denisovans and Homo sapiens must have occurred in Asia, at some time in the many millennia during which humans migrated from Africa to Asia and then to Australia and Melanesia.
Question 8.15
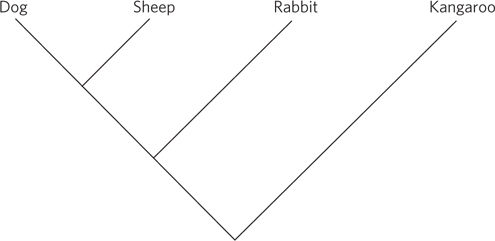
(a) Coronavirus genomes are composed of a single strand of RNA. At least one round of synthesis by reverse transcriptase is needed if the genome is to be amplified by PCR. (b) In all PCR experiments, researchers design primers that are unique and sufficiently long that they are unlikely to anneal well to other areas of a genome. In an experiment like this, one would also search for sequences that are highly conserved among similar genomes (e.g., sequences encoding parts of an enzyme that are critical to the enzyme’s function), thereby maximizing the chance that the sequence will be present in an unchanged state in the target genome. (c) HEV and BCoV: 6; BCoV and SARS: 146; TGEV and SARS: 168. The coronaviruses BCoV and HEV are most closely related, with few differences between them. The differences among sequences are consistent with the phylogenetic tree.