Histone H1 Binds the Nucleosome
Treating chromatin with a nonspecific nuclease to digest all naked DNA (DNA not protected by protein) yields a segment of about 168 bp to which all five histones are bound—the four core histones plus histone H1. More extensive nuclease digestion releases H1, leaving the core histone octamer bound to about 147 bp of DNA (i.e., the nucleosome). Indeed, the addition of H1 to a nucleosome results in protection of an additional 20 to 22 bp of linker DNA adjacent to the nucleosome, and thus H1 is often referred to as the linker histone. Only one H1 subunit is present per histone octamer, unlike the core histones, which are present in two copies each.
Compared with the core histones, H1 is more variable in sequence. Most organisms even have multiple H1 subtypes. For example, mammals have eight H1 variants that differ in their ability to condense chromatin. The H1 subtypes are expressed at different times of development or are present in different cell types. The avian counterpart of H1 is known as H5.
H1 consists of a short, 20 to 35 residue N-terminal region, a central globular domain of about 80 amino acid residues, and a long C-terminal region of about 100 residues. DNA binding is intrinsic to the central globular region, which contains two DNA-binding sites. It was originally thought that the two DNA-binding sites in H1 were used to bind each of the two linker DNA strands at the sites where DNA enters and exits the nucleosome. However, more recent studies indicate that H1 binds only one of the linker DNA strands, and the second DNA-binding site in histone H1 binds to the central region of the DNA supercoil in the nucleosome (Figure 10-11).
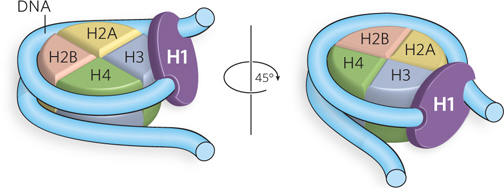
Figure 10-11: The binding of DNA by histone H1. Two views of the nucleosome containing histone H1. H1 has two DNA-binding sites, through which it makes contact with one arm of linker DNA and the central region of the DNA wrapped around the histone octamer.
By binding an additional 20 bp of DNA, histone H1 alters the DNA entry and exit angles, facilitating the packing of DNA into higher-order chromatin structures (Figure 10-12). When H1 is extracted from chromatin, DNA seems to enter and exit nucleosomes at different places, thus creating the beads-on-a-string appearance (see Figure 10-3b). The presence of H1 causes DNA to enter and exit the nucleosome at nearly the same place, resulting in a zigzag pattern with a DNA entry/exit angle between 40° and 100°, depending on the conditions of sample preparation. Overall, the level of condensation provided by H1 is six to seven times that of the nucleosome, for a total length reduction of 35- to 40-fold in chromatin. H1 helps nucleosomes condense into a higher level of packaging, the 30 nm filament (described below).
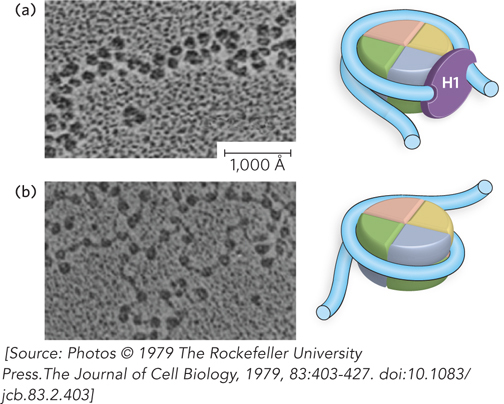
Figure 10-12: The zigzag appearance of nucleosomes in the presence of histone H1. Electron micrographs of nucleosomes (a) in the presence of H1 and (b) in the absence of H1. Histone H1 increases the zigzag appearance by decreasing the DNA entry/exit angles, as shown in the cartoon models to the right of each micrograph. In these electron micrographs, the samples were stained by rotary shadowing using carbon-platinum. Scale bar = 1,000 Å.
Nucleosomes generally repress transcription by sterically preventing the access of regulatory proteins to promoter sequences. H1 participates in this activity by stabilizing nucleosomes on the DNA and promoting higher-order chromatin structure that further compacts nucleosomal DNA. Indeed, most regions of actively transcribed DNA are known to lack histone H1.
Chromosomes Condense into a Compact Chromatin Filament
Under certain experimental conditions, such as increased ionic strength or the presence of particular divalent cations, nucleosomes condense into a compact filament with a width of about 30 nm, referred to as the 30 nm filament (Figure 10-13a). The 30 nm filament is thought to exist in living cells, but this has yet to be rigorously proven. Although histone H1 promotes condensation into the 30 nm filament, it is not essential for forming it. In contrast, the N-terminal tails of the core histones are absolutely required, suggesting that the tails provide important nucleosome-nucleosome contacts needed for 30 nm filament formation. Recall that the crystal structure of the nucleosome shows that the terminal tails of H4, H3, and H2A make contact with adjacent nucleosomes, and these contacts may be involved in the condensation of nucleosomes into a filament.
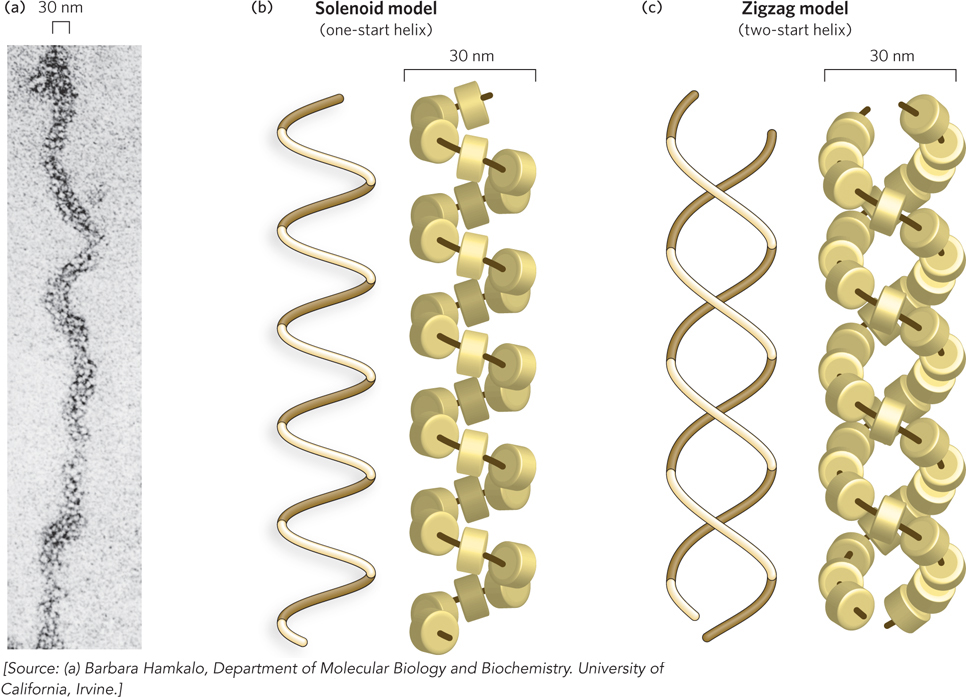
Figure 10-13: The 30 nm filament, a higher-order organization of nucleosomes. The compact filament is formed by the tight packing of nucleosomes. (a) The 30 nm filament as seen by electron microscopy. Two proposed models of filament structure are (b) the solenoid model and (c) the zigzag model. The path of DNA is shown as a dark brown line through the histones, for clarity of histone visualization.
The exact arrangement of nucleosomes in a 30 nm filament is still unclear, although any model should accommodate the following observations: (1) neutron diffraction studies place histone H1 in the center of the filament; (2) linker DNA should also be placed in the center of the filament, because H1 binds linker DNA; and (3) electron microscopy and x-ray diffraction studies indicate that the nucleosome units in the 30 nm filament are arranged with a helical pitch of about 11 nm, the width of a nucleosome.
The two most widely accepted models for nucleosome arrangement in the 30 nm filament fit these criteria, and both are supported by substantial experimental evidence. The two models may in fact be alternative ways in which nucleosomes pack in different areas of the same 30 nm filament. In the solenoid model (also called the one-start helix model), the nucleosome array adopts a spiral shape, in which the flat sides of adjacent nucleosome disks are next to each other (Figure 10-13b). The linker DNA is presumed to bend inside the center of the filament to account for the observed constant thickness of the fiber with different linker lengths.
The second model for the 30 nm filament is the zigzag model (also called the two-start helix model), in which zigzag histone pairs stack on each other and twist about a central axis (Figure 10-13c). The zigzag model was inspired by the appearance of nucleosomes under the electron microscope (see Figure 10-12). This model is supported by the crystal structure of a tetranucleosome, in which four nucleosomes are bound to one DNA molecule (Figure 10-14a). The structure shows two histone-pair zigzags stacked on top of each other, with the linker DNA passing through the central axis connecting two nucleosomes on opposite sides of the filament. A zigzag model of a chromatin filament of many nucleosomes, based on the tetranucleosome structure, is shown in Figure 10-14b and c.
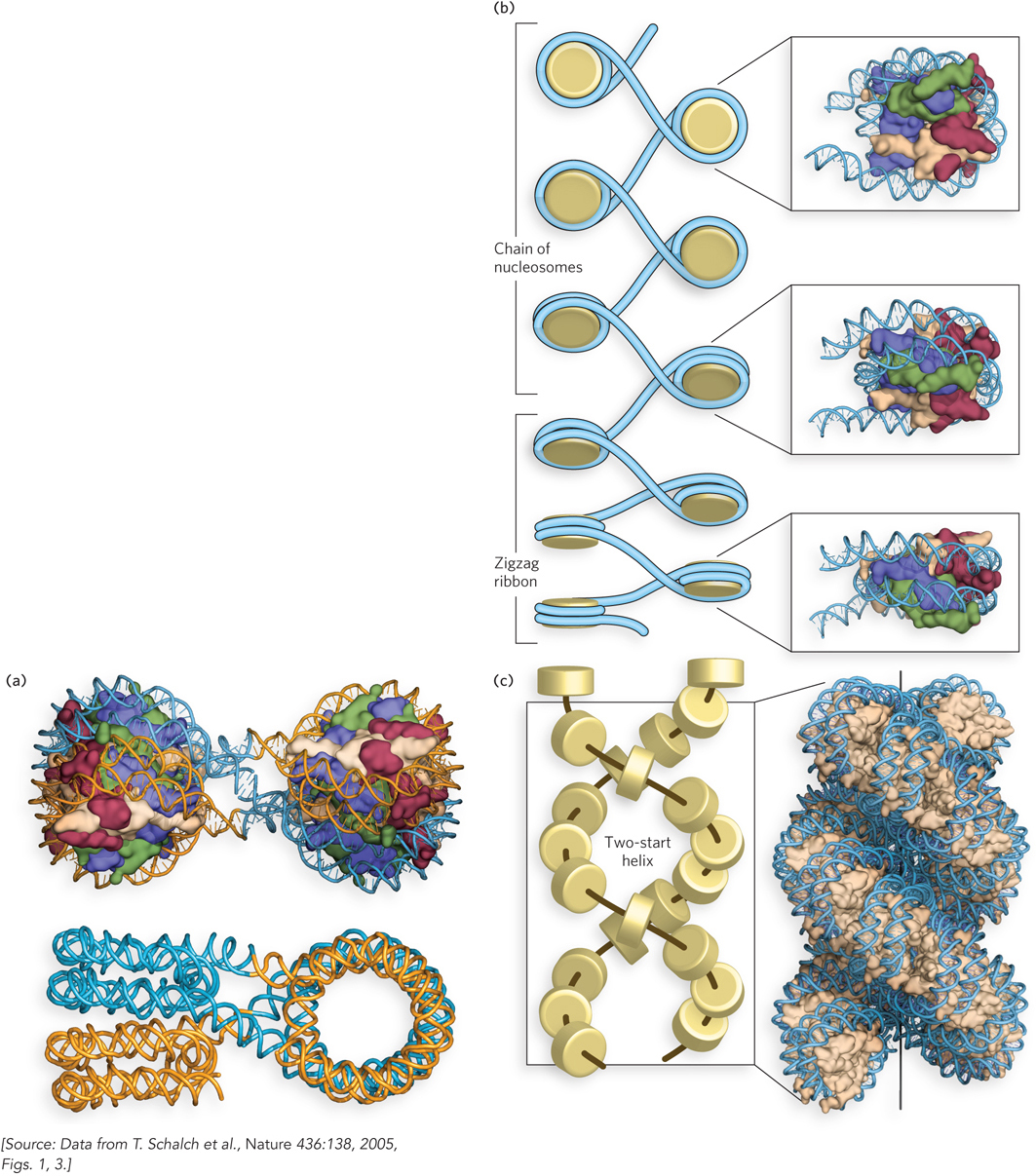
Figure 10-14: The crystal structure of a tetranucleosome. (a) The four nucleosomes are arranged in two zigzag pairs that stack on top of each other. Shown here are the DNA and histone octamers (top) and the DNA only (bottom), in orientations that illustrate the geometry of the two stacks. (b and c) A model of the 30 nm filament made by repeating the tetranucleosome in a continuous zigzag configuration.
The two models for nucleosome stacking in a 30 nm filament are fundamentally very different, so it might seem they would be easy to distinguish experimentally. The inability of investigators to resolve this issue reflects the many irregularities in natural chromatin fibers, leading to the poor quality of experimental data. However, as noted above, both types of nucleosome packing might occur in the same chromatin filament.
Higher-Order Chromosome Structure Involves Loops and Coils
DNA is much more condensed inside chromosomes than in the 30 nm filament. Treating chromosomes with a low-salt buffer causes them to expand, and the edges of these swollen chromosomes reveal 30 nm filaments that appear to be organized in loops estimated at 40 to 100 kbp long (Figure 10-15a). The existence of loops of DNA as a substructure within chromosomes is also supported by electron micrographs of histone-depleted chromosomes. Histones can be selectively extracted from chromosomes by treatment with negatively charged polymeric chemicals, such as heparin and dextran sulfate, which compete with the DNA for binding histones. After histone extraction, a proteinaceous residue remains that retains the size and shape of the original chromosome. This residue is called the chromosomal scaffold (Figure 10-15b). One of the major components of the chromosomal scaffold is the SMC proteins that hold DNA strands together, keeping eukaryotic chromosomes topologically constrained (see Figure 9-11). The histone-depleted DNA can be seen to form large loops anchored in the scaffold (Figure 10-15c), which is consistent with looping as a higher-order arrangement of DNA in chromosomes.
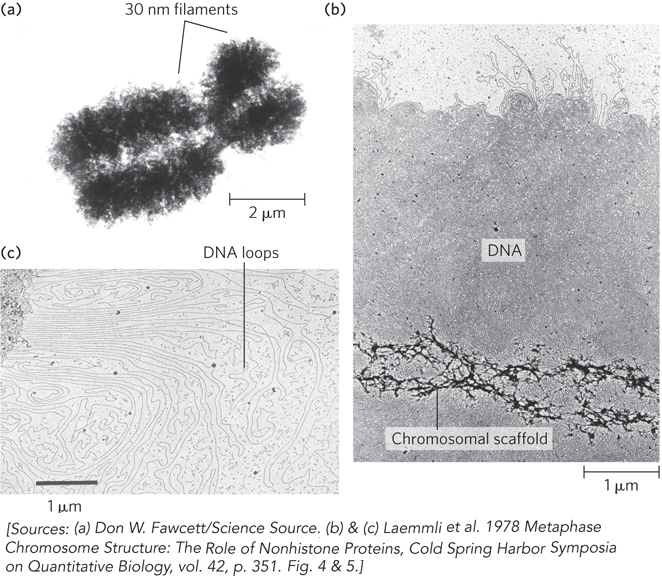
Figure 10-15: Loops of DNA attached to a chromosomal scaffold. (a) A swollen chromosome, produced in a buffer of low ionic strength, as seen in the electron microscope. Notice the appearance of 30 nm filaments (chromatin loops) at the margins. (b) Extraction of the histones leaves a proteinaceous chromosomal scaffold surrounded by naked DNA. (c) The DNA appears to be organized in loops attached at their base to the scaffold in the upper left corner. Note the different magnifications for the three images.
Although very little is known about further steps of DNA condensation beyond the 30 nm filament, there are probably several more layers of organization in eukaryotic chromosomes, each increasing the degree of compaction. Many speculative models have been proposed, one of which is shown in Figure 10-16. In this model, loops of the 30 nm filament are connected to the scaffold in a radial fashion. One radial turn forms a rosette composed of six loops, held together by SMC proteins (see Figure 9-27). A spiral of 30 rosettes per turn forms a coil, and a chromosome consists of several coils.
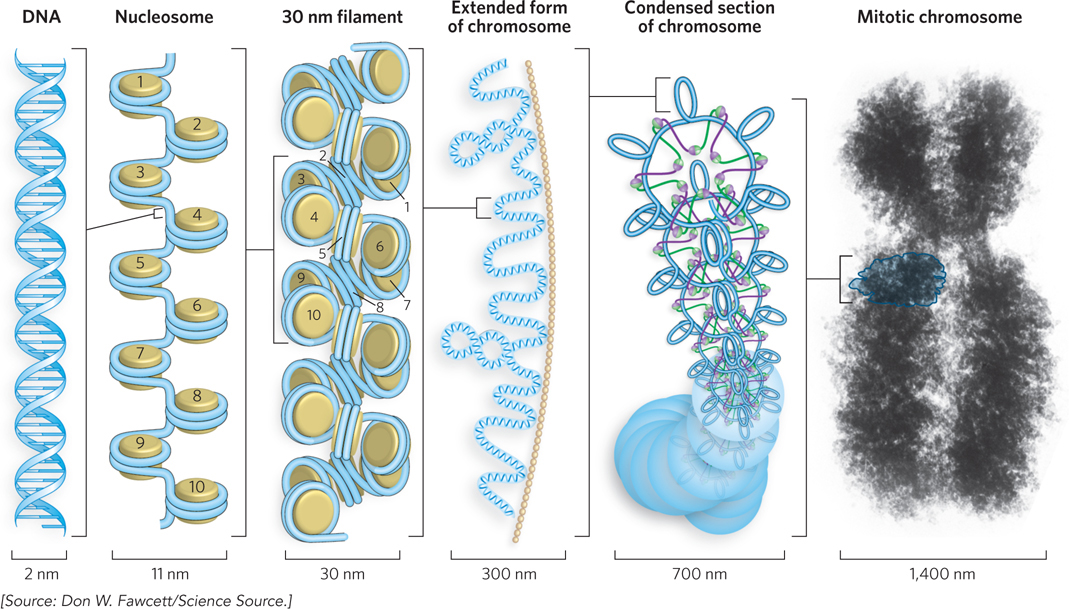
Figure 10-16: Higher-order DNA compaction in a eukaryotic chromosome. This model shows the levels of organization that could provide the observed degree of DNA compaction in the chromosomes of eukaryotes. First the DNA is wrapped around histone octamers, then H1 stimulates formation of the 30 nm filament (solenoid model is shown). Higher levels of organization are not well understood but seem to involve further coiling and loops in the form of rosettes, which also coil into thicker structures. Overall, progressive levels of organization take the form of coils upon coils upon coils. In cells, the higher-order structures (above the 30 nm filament) are unlikely to be as uniform as depicted here.
In reality, the higher-order structure of chromatin probably varies from chromosome to chromosome, from one region to the next in a single chromosome, and from moment to moment in the life of a cell. No single model can adequately describe these structures. Nevertheless, the principle is clear: DNA condensation in eukaryotic chromosomes probably involves coils upon coils upon coils.
Bacterial DNA, Like Eukaryotic DNA, Is Highly Organized
Bacterial DNA is compacted in a structure called the nucleoid, which can occupy a significant fraction of the cell’s volume (Figure 10-17a). The DNA seems to be attached at one or more points to the inner surface of the cytoplasmic (plasma) membrane. Much less is known about the structure of the nucleoid than of eukaryotic chromatin. Bacteria contain SMC proteins, and studies in E. coli reveal a scaffoldlike structure that seems to organize the circular chromosome into a series of about 500 looped domains, each encompassing 10 kbp, on average (Figure 10-17b). Like the looped domains in eukaryotic chromosomes, the looped DNA domains in the bacterial chromosome are topologically constrained. For example, if the DNA is cleaved in one domain, only the DNA in that domain becomes relaxed. However, bacterial DNA does not seem to have any structure comparable to the local organization provided by nucleosomes in eukaryotes.
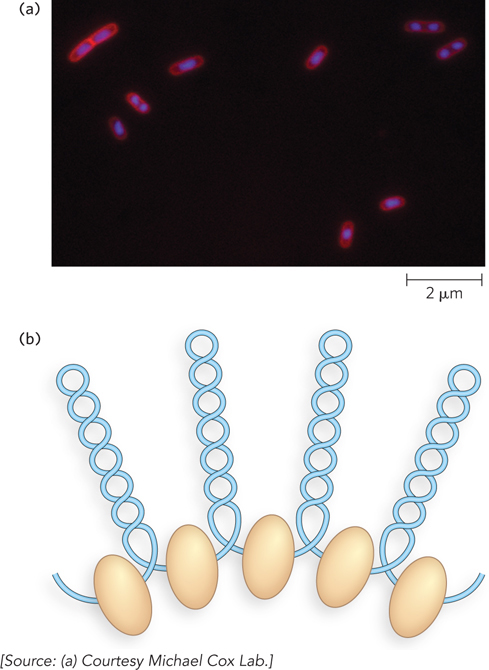
Figure 10-17: Highly condensed bacterial DNA. (a) The DNA is stained with a dye that fluoresces when exposed to UV light. The light areas define the nucleoids. Notice that some cells have replicated their DNA but have not yet undergone cell division and hence have multiple nucleoids. (b) Looped domains of DNA in the bacterial chromosome are separated by points of connection to the scaffold, shown as beige ovoids.
Although bacteria lack nucleosomes, they do contain abundant histonelike proteins. A well-studied example is the two-subunit protein HU (Mr 19,000). Bacterial histonelike proteins do not seem to form stable oligomeric structures, and this may reflect the need for bacteria to respond very rapidly to their environment, requiring more ready access to their genetic information. For example, bacterial cell division can be as short as 15 minutes, whereas a typical eukaryotic cell may not divide for hours or even months. In addition, a much greater proportion of bacterial DNA than eukaryotic DNA is used to encode protein or functional RNA. Furthermore, higher rates of cellular metabolism in bacteria mean that a much larger proportion of their DNA is being transcribed or replicated at a given time than in most eukaryotic cells.
SECTION 10.2 SUMMARY
The histone octamer and associated DNA that form the nucleosome combine with histone H1. H1 binds additional DNA, altering the entry and exit angles of the DNA so that the nucleosomes pack in a zigzag pattern.
Two models describe how nucleosomes might pack into a 30 nm filament. In the solenoid model, nucleosome disks are next to one another, forming a spiral shape. In the zigzag model, pairs of nucleosomes stack on top of each other and twist about a central axis.
Higher-order chromatin structure is largely undetermined, but most likely involves loops that form topologically constrained domains. Looped domains form rosettes, which form coils. Chromosome shape is determined by a rigid proteinaceous scaffolding containing SMC proteins, among other components.
Bacteria lack nucleosomes, but their chromosomal DNA is compacted into looped domains.






