13.1 RECOMBINATION AS A DNA REPAIR PROCESS
As discussed in Chapter 12, DNA damage is common and highly deleterious. The most important consequences of the damage do not become apparent until the DNA is replicated. When a lesion exists in the DNA template, one of several things can happen: replication continues over the lesion, leaving it in place; lesion repair is initiated but not completed, such that a break in the template strand is present when the replication fork arrives; replication stalls, requiring repair before it can continue; or replication stalls but picks up again at a location downstream of the lesion.
In the first possible outcome, DNA synthesis continues over the lesion by translesion synthesis (TLS), a process described in Chapter 12 (Figure 13-1a; see also Figure 12-26). Normally, this occurs only with the aid of specialized DNA polymerases capable of TLS. Translesion DNA polymerases are found in all cells and are often adapted to particular repair scenarios. More rarely, the usual DNA-replicating polymerase may replicate over lesions that do not cause significant DNA distortion. For example, an O6-meG in the template strand pairs with T rather than C, and replication of the lesion will result in a C→T transition mutation.
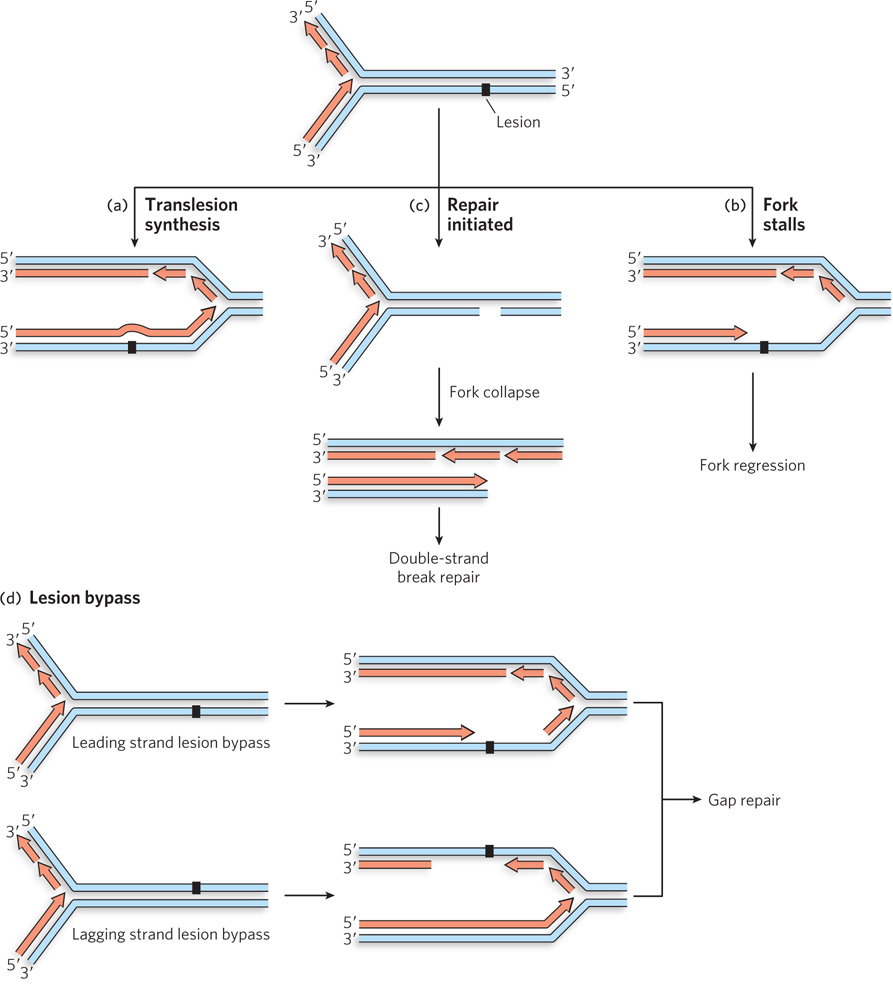
Figure 13-1: Possible effects of a damaged template on a replication fork. (a) Translesion synthesis occurs when the replisome encounters a lesion and keeps going. (b) If the replisome encounters a lesion that is undergoing repair, a single-strand break can cause the replication fork to collapse, creating a double-strand break. (c) Some lesions prevent progress of the replisome, resulting in a stalled replication fork. (d) In some cases, the lesion is bypassed, leaving a single-strand gap, and replication continues downstream.
If the replisome encounters a lesion that is already undergoing nucleotide excision repair (NER) or base excision repair (BER), the template strand may contain a temporary break. When the replication fork arrives, one branch of the fork becomes disconnected and the replication fork collapses (Figure 13-1b)—a particularly catastrophic outcome that creates a double-strand break. Here, recombinational DNA repair restores an undamaged fork structure, allowing replication to restart.
Most lesions, if encountered before any repair process is initiated, cause the replication fork to stall (Figure 13-1c). The replisome cannot insert a nucleotide opposite the lesion and replication ceases until the lesion is repaired.
In still other cases, the replication machinery is blocked by the lesion but resumes replication further downstream (Figure 13-1d). The lesion is left behind in a single-strand gap, with no undamaged complementary strand present to guide the most common DNA repair pathways. This kind of bypass-plus-restart outcome occurs most readily when the lesion is in the lagging strand, because the inherent mechanism used to initiate the synthesis of new Okazaki fragments can simply continue downstream.
Recombinational DNA repair resolves each of the situations illustrated in Figure 13-1b–d, although the pathways differ in ways large and small, and in some cases even use somewhat different sets of enzymes. As described more fully below, a stalled replication fork is repaired by fork regression, a collapsed fork with a DSB is resolved by DSB repair, and the gap resulting from lesion bypass is filled by DNA gap repair.
Double-Strand Breaks Are Repaired by Recombination
Double-strand breaks can result from oxidative DNA damage. This is a relatively rare occurrence, but can be a byproduct of respiration in organisms growing in an oxygen-rich environment or a consequence of exposure to ionizing radiation. These lesions destroy the continuity of both template strands, and they are generally lethal if not repaired. More commonly, DSBs arise when the replication fork encounters a break in one template strand that is undergoing repair (see Figure 13-1b). We first consider a generalized pathway for the repair of DSBs—a pathway that will reappear in slightly different forms as we discuss specific repair pathways. The enzymes we encounter are described in more detail in Section 13.2.
The repair of double-strand breaks by recombinational DNA repair requires the presence of another, undamaged, homologous double-stranded DNA. In a diploid cell, that double-stranded DNA is either the second copy of each chromosome or the sister chromatid present immediately after DNA replication. This second DNA molecule guides the repair process by providing a template for the restoration of genetic information that might otherwise be lost as nucleotides missing at the site of the break.
We illustrate the process by examining the repair of a DSB in a chromosome present in a diploid eukaryotic cell. The pathway for recombinational DNA repair of the DSB is initiated by three enzymatic reactions that define every process that makes use of homologous recombination. First, the broken DNA ends are processed, with the 5′-ending strands selectively degraded to create 3′ single-stranded extensions, or overhangs, at the site of the break (Figure 13-2, step 1). Second, the 3′ single-stranded extensions invade the homologous chromosome, in a process catalyzed by a ubiquitous class of enzymes called recombinases. This DNA strand invasion is the quintessential step of homologous recombination and all processes related to it. The invading strand displaces one strand of the intact homologous chromosome and base-pairs with the other. The structure created by strand invasion by the 3′ single-stranded extension is sometimes referred to as a D-loop. In the pathway shown in Figure 13-2, two consecutive strand invasions are shown in steps 2 and 3. The third defining reaction is the replicative extension of the invading strand. The use of 3′ ends for the invasion step has the important consequence that these ends can also act as primers for DNA synthesis. DNA polymerase–mediated extension of the invading strands after they are paired (step 4) lengthens them in a manner that faithfully restores any information lost at the site of the break, using the invaded chromosome as the template. For the pathway shown in Figure 13-2, the two strand invasions produce two DNA crossover points, where strands originating separately from the two participating DNA molecules are base-paired to each other.
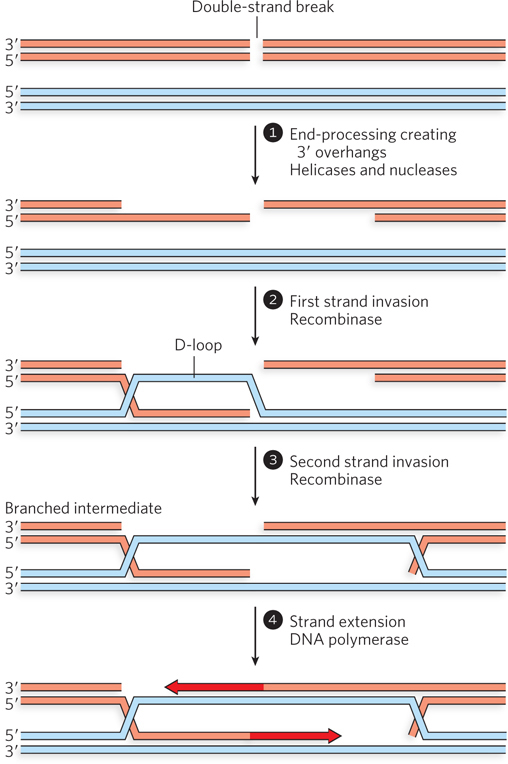
Figure 13-2: The repair of chromosomal double-strand breaks. Accurate repair of a DSB requires an undamaged source of duplicate genetic information—that is, homologous double-stranded DNA. The broken ends are first processed to generate 3′ single-stranded extensions, and both ends are used for strand invasion of the homologous double-stranded DNA to form D-loops. The invading 3′ ends are extended by DNA polymerases.
Several additional steps complete the repair process. The DNA double-crossover intermediate seems complex, but virtually every cell can resolve it in at least two ways. First, the now-lengthened invading strands can simply be displaced by the action of helicases and then anneal to each other (Figure 13-3a). Any remaining gaps can be filled by DNA polymerases. DNA ligases complete the repair by ligating the ends. This quite common pathway is known as synthesis-dependent strand annealing (SDSA).
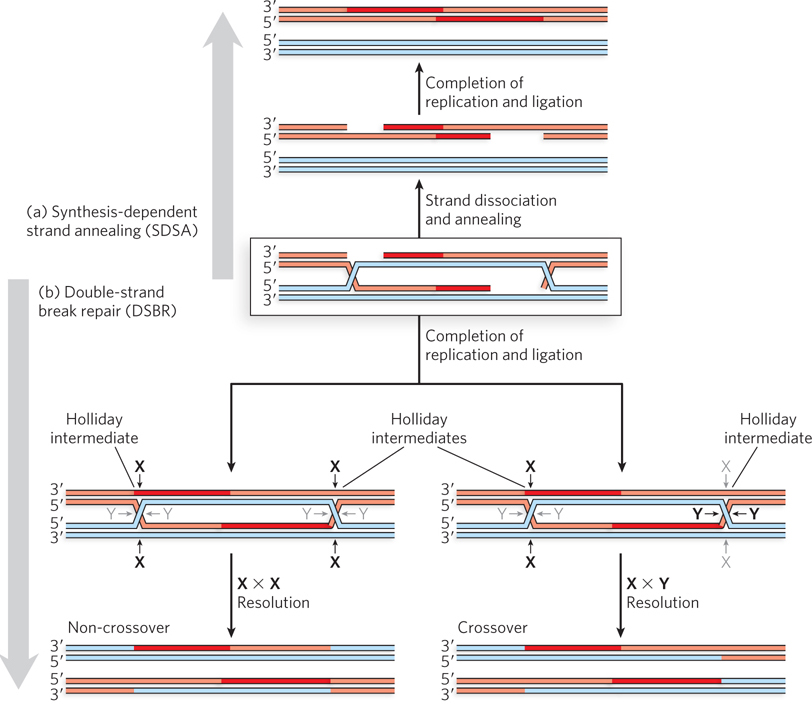
Figure 13-3: Two paths for completing double-strand break repair. (a) In the SDSA pathway, the invading strands dissociate and anneal to each other. Further replication and ligation complete the process. (b) In the DSBR pathway, the two Holliday intermediates are cleaved by Holliday intermediate resolvases (see Section 13.2). Resolution of the Holliday intermediates can yield two different results with respect to the DNA flanking the repair site, as described in the text.

Robin Holliday, 1932–2014
In the second possible pathway (Figure 13-3b), double-strand break repair (DSBR), replication is completed by ligating the strands while they are still linked. A four-branched crossover junction, with all DNA strands intact such that each branch is a segment of duplex DNA, is called a Holliday intermediate (or Holliday junction)—named for the geneticist Robin Holliday, who proposed the first recombination model that included these structures. Two Holliday junctions are generated in the pathway shown in Figure 13-3b. Specialized endonucleases present in all cells, called Holliday junction resolvases, recognize and cleave the Holliday intermediates in one of two ways. In both, cleavage by the resolvases creates products that are viable chromosomes with a complete set of genes. The breaks left behind by the cleavage are sealed by DNA ligase. The cleavage and rejoining may result either in the retention of the chromosomal DNA segments originally linked on either side of the repair site or in an exchange of that information, resulting in a genetic crossover. In Figure 3-13b, the former result occurs if both Holliday intermediates are cleaved at the sites labeled X or both are cleaved at the sites labeled Y. A genetic crossover occurs if one Holliday junction is cleaved at the X sites and the other at the Y sites, such that the genetic material extending from the site of repair to the telomere is transferred between chromosomes.
In principle, recombination can occur between any two DNA segments that share sequence similarity. Thus, deleterious events may occur. For example, recombination of two repeated sequences on the same chromosome could lead to complete deletion of all the genetic information between these sequences. Such deleterious events are rare, given the very tight regulation imposed on homologous recombination systems in all cells.
With this overview in mind, we now look at how the various steps of SDSA and DSBR are applied to the repair of replication forks. When replication forks stall or collapse, recombinational DNA repair has a major advantage over translesion DNA synthesis: it does not cause mutations.
Collapsed Replication Forks Are Reconstructed by Double-Strand Break Repair
When a replication fork encounters a break in a template strand, one arm becomes detached (see Figure 13-1b). The resulting double-strand break and collapse of the fork triggers a recombinational repair process. Here, and in some other pathways we will soon encounter, the steps shown in Figure 13-2 will become quite familiar. In brief, repair requires the reattachment of the broken arm to recreate the fork. The reattachment involves processing of the broken DNA end to create a 3′ single-stranded extension. A DNA strand invasion reaction is then mediated by a recombinase.
The reconstruction of a replication fork by recombinational DSBR is shown in Figure 13-4a. The broken DNA end is processed by nucleases to remove a segment of the strand with the free 5′ end at the break (step 1). This creates the 3′ single-stranded DNA extension. A recombinase binds to the single-stranded DNA and promotes a strand invasion of the intact portion of the chromosome, so that the single-stranded extension is paired with its complementary strand (step 2). At the point of insertion, the other strand of the invaded duplex DNA is displaced. Once the strand invasion is complete, the repair of a replication fork often diverges from the path shown in Figure 13-2 in that the invading strand is not extended. Instead, reconstruction of the replication fork requires the action of enzymes that promote a process called branch migration (step 3; Figure 13-4b shows the details of this process). A DNA branch is formed when separate segments of at least one DNA strand are paired with two different partner strands. In branch migration, DNA branches are moved along the DNA, with some base pairs forming and others being disrupted, but with no net increase or decrease in number of paired nucleotides. Branch migration can occur spontaneously in branched DNA molecules and takes place through a kind of random walk mechanism, with movement occurring in either direction. In a cell, branch migration is usually catalyzed such that it moves in one direction only. Branch migration may create a Holliday intermediate, which is resolved by the specialized Holliday intermediate resolvases, along with DNA ligase (see Figure 13-3a, step 4). The reconstructed fork is then ready for renewed replication, and no mutations have been added in the process. Replication is restarted with the aid of a dedicated replication restart complex. In bacteria, the replication restart enzymes reload the replicative DnaB helicase onto the DNA. The other replisome components load spontaneously onto DnaB, and replication begins anew.
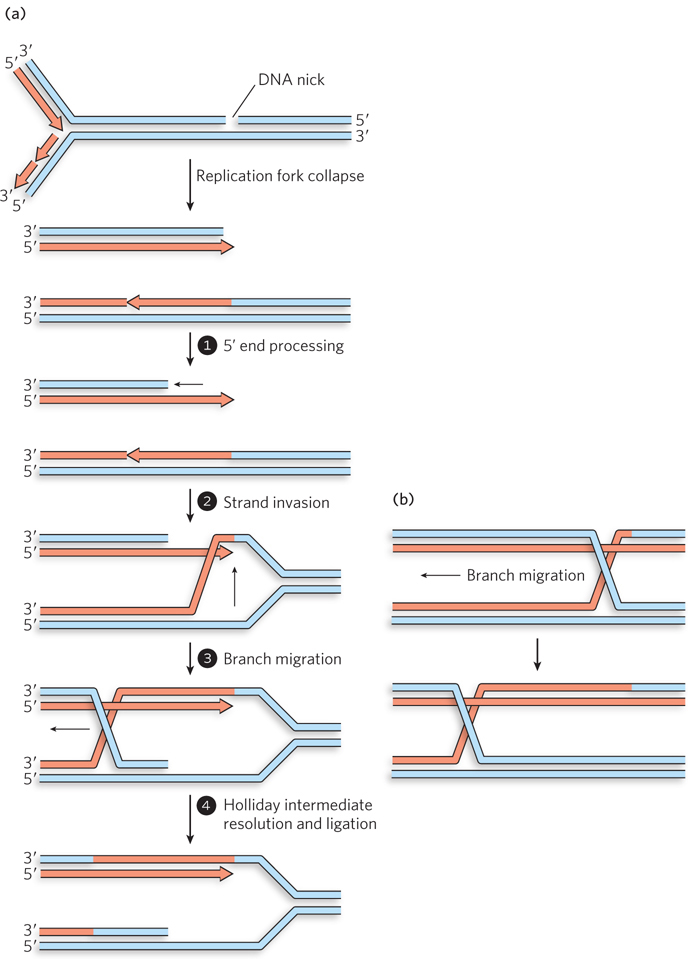
Figure 13-4: Recombinational DNA repair at a collapsed replication fork. (a) When a replication fork encounters a template strand break, one arm of the fork is lost and the fork collapses. The 5′ end at the break is processed to create a 3′ single-stranded extension, which is used in a strand invasion reaction. Migration of the branch can create a Holliday intermediate. Resolution of the Holliday intermediate, followed by ligation, restores a viable replication fork. The replisome is reloaded onto this restored fork (not shown), and replication continues. (b) Details of branch migration. In this process, the branch moves, but the net amount of duplex DNA does not change. Base pairs separated on one side of the branch are replaced on the other side.
A Stalled Replication Fork Requires Fork Regression
When the replication fork encounters a template strand lesion that cannot be surmounted, the fork stalls at the lesion. In some cases, replication restarts downstream; in others, replication simply halts until the lesion is repaired. In either case, the lesion is left without a complementary strand. Most of the DNA repair processes described in Chapter 12—nucleotide excision repair, base excision repair, and mismatch repair—require that the lesion occur in only one strand of a duplex DNA. In these repair mechanisms the lesion is simply removed from the damaged strand, and the undamaged complementary strand guides the replication and ligation needed to fill in the missing nucleotide(s). For a lesion left in a single-strand gap, the reactions shown in Figure 13-5 are directed at providing a complementary strand to allow normal repair of the lesion and to complete the process without creating a mutation.
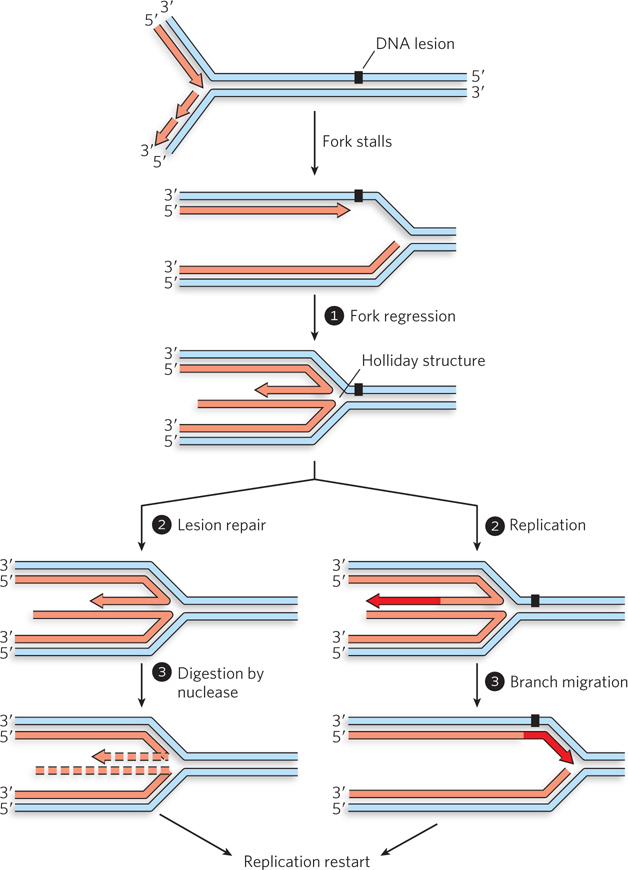
Figure 13-5: Repair of a replication fork stalled at a DNA lesion. When a replication fork encounters a blocking lesion in the leading strand, the fork stalls. Fork regression creates a Holliday intermediate and reunites the lesion-containing strand with the complementary template strand with which it was originally paired. Lesion repair can then proceed by the usual pathways. Replication can be restarted by digestion of the short arm and reloading of the replisome (steps 2 and 3, left). Alternatively, the 3′ end of the short arm can be extended to the end of the available template (step 2, right). If the lesion is not repaired, branch migration in the direction opposite to fork regression pairs the newly synthesized strand with the lesion (step 3, right). Replication is restarted, and repair can occur later.
In many cases, branch migration forces the fork backward, a process known as fork regression (see Figure 13-5, step 1). As the fork migrates in reverse, the lesion-containing strand is reunited with its original complementary strand, and the newly synthesized DNA strands are paired with each other. This results in a four-branched Holliday intermediate at the point where the four strands intersect. The regression leaves the lesion paired with its undamaged complementary strand, allowing its repair by processes such as NER (see Chapter 12). If the lesion is repaired, the Holliday intermediate can be resolved by exonucleolytic degradation of the short DNA arm, as shown in Figure 13-5 (steps 2 and 3, left). A second method of resolving the Holliday intermediate can occur whether or not the lesion has been repaired. This method entails replication of the short DNA arm, followed by branch migration in the opposite direction (steps 2 and 3, right). Even if the lesion has not been repaired at this point, it is now paired with a complementary DNA strand that can be used as a template for later repair. In either case, replication must be restarted.
Single-Stranded DNA Regions Are Filled In by Gap Repair
In some cases, DNA polymerase bypasses a DNA lesion by halting and then continuing DNA synthesis upstream. This leaves the lesion within a single-strand gap, where it is not readily repaired. The repair pathway must then generate an undamaged complementary strand. The gap can be filled in by translesion synthesis (see Figure 12-26), but TLS can be mutagenic and is usually not the pathway of first choice. Alternatively, recombinational DNA repair can again be used, in a variant called gap repair (Figure 13-6). In this mechanism, the complementary strand needed for repair of the lesion is found in the undamaged arm of the replication fork.
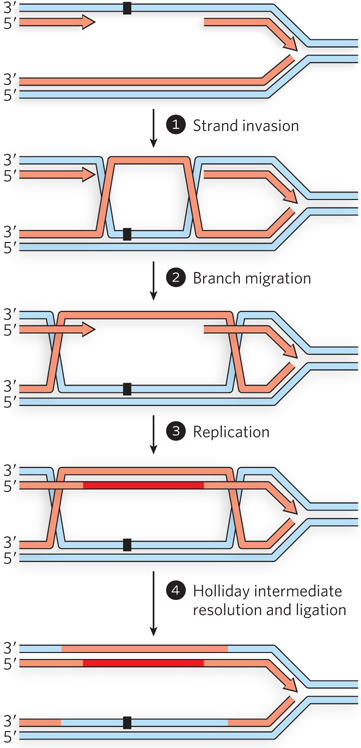
Figure 13-6: Repair of a DNA gap after the replication fork bypasses a lesion. If a single-strand gap is left behind by the replication fork, the information for its repair must come from the other side of the fork. A recombinase-mediated strand switch provides an undamaged template strand, which is subsequently used to extend the discontinuous strand in the gap. The process is completed by resolution of the Holliday intermediates and ligation.
The steps shown in Figure 13-6 follow a pattern that should now be familiar, with some subtle variations. A recombinase binds to the DNA in the single-strand gap. The recombinase promotes a strand invasion, using the bound single-stranded DNA to invade the undamaged double-stranded DNA on the other side of the replication fork (step 1). In this instance, the invading DNA has no free end. Branch migration (step 2) generates two Holliday intermediates. The displaced strand serves as an undamaged template for replication (step 3). Both Holliday intermediates are then cleaved by Holliday intermediate resolvases, and the nicks are sealed by DNA ligase (step 4). The resolution can result in a crossover; however, many cells have enzymatic systems that direct most resolution events into a non-crossover pathway. In the end, the strand that had the DNA lesion now has an undamaged complement, and nonmutagenic repair by NER or BER can begin to remove the lesion.
The pathways described in Figures 13-4, 13-5, and 13-6 provide responses to almost any deleterious event that can befall a replication fork. Recombinases, acting in concert with DNA polymerases, ligases, Holliday intermediate resolvases, and proteins that promote branch migration, provide a repair system that readily adapts to the variety of stalled or collapsed fork structures that may present themselves. In the next section, we consider these enzymes in more detail.
SECTION 13.1 SUMMARY
Recombination is a pathway for the repair of single-strand and double-strand breaks, both of which can arise during replication.
Double-strand break repair entails the generation of 3′ single-stranded extensions at the broken DNA ends, which are then paired with a homologous duplex region in a strand invasion reaction catalyzed by recombinases.
Many recombination reactions produce a four-branched DNA intermediate called a Holliday intermediate.
Recombinational DNA repair restores DNA segments with double-strand breaks to their original genomic condition, causing no mutations.
When a replication fork encounters a single-strand break in the template DNA, the break is converted to a double-strand break, and the replication fork collapses. The fork is then repaired by double-strand break repair, and replication is restarted.
When a replication fork encounters a nucleotide lesion, it may bypass the lesion by translesion synthesis or it may stall. Stalled forks may be repaired by a pathway involving fork regression.
When lesions are left in single-strand gaps, recombination provides a nonmutagenic pathway for gap repair.






