PROBLEMS
Question 14.1
Holliday intermediates are associated with both homologous genetic recombination and some site-
Question 14.2
Compare and contrast the DNA sequence requirements of homologous genetic recombination, site-
Question 14.3
If two FRT sites are present in a single DNA molecule in the same orientation, the Flp recombinase will promote a deletion of the DNA between the two sites. If the FRT sites are altered so that their core regions have a perfectly symmetric (palindromic) sequence instead of the usual asymmetric sequence, how might the fate of the intervening DNA in the reaction change? (Ignore any intermolecular reactions between the DNA molecules.)
Question 14.4
When a site-
Question 14.5
Draw the products of Cre-

Question 14.6
What are the possible products of Flp recombinase–
516
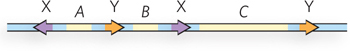
Question 14.7
Which of the following DNA molecules are appropriate substrates for the Hin recombinase? Arrowheads indicate the location and orientation of hix sites.
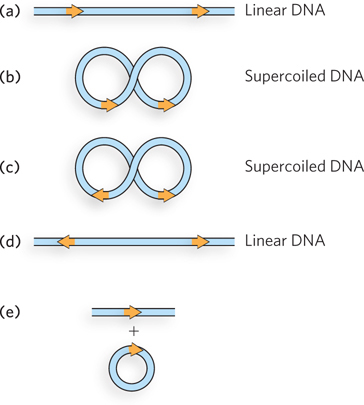
Question 14.8
The bacterial transposon Tn3 uses a replicative transposition pathway to move from one DNA molecule to another. Tn3 encodes not only a transposase but a site-
Question 14.9
The Flp and Cre-
Question 14.10
Cut-

Question 14.11
Many retroviruses have sequences complementary to the 3′ end of one of the tRNA molecules prominent in host cells infected by the virus. What is the purpose of this complementarity?
Question 14.12
The transposase from the bacterial transposon Tn5 initiates transposition by creating a double-
Question 14.13
Many TP (non-
Question 14.14
The human genome has more than a million copies of the SINE element Alu. These transposons are found in the DNA between genes, and often in the introns of genes, but very rarely in gene exons. Explain why this is so.