DATA ANALYSIS PROBLEM
Cech, T.R., A.J. Zaug, and P.J. Grabowski. 1981. In vitro splicing of the ribosomal-
Kruger, K., P.J. Grabowski, A.J. Zaug, J. Sands, D.E. Gottschling, and T.R. Cech. 1982. Self-
Question 16.18
The discovery of RNA catalysis is sometimes cited as a classic case of serendipity, but it required a thoroughly prepared mind. In the late 1970s and early 1980s, RNA splicing was still a new concept. Thomas Cech and his colleagues set out to investigate the splicing of an intron in rRNA of the protozoan Tetrahymena thermophila. They selected this RNA for several reasons: rRNAs are much more abundant than most mRNAs, this rRNA has an intron, and Tetrahymena is a single-
To carry out their early studies, Cech and colleagues devised a method to produce unspliced precursor rRNAs. As described in their 1981 paper, they isolated nuclei from Tetrahymena cells, lysed them, and added buffer, labeled rNTPs, and α-amanitin, then incubated the reaction mix under conditions in which the cellular RNA polymerases would synthesize RNA. They extracted the labeled RNA by a method that used phenol and chloroform, treatments that normally remove most protein.
Why did the researchers add α-amanitin to the transcription reaction?
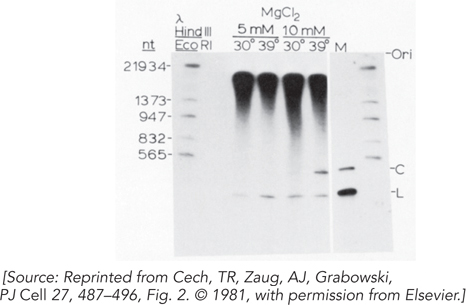
When they incubated the extracted RNA under the right conditions, the researchers saw the production of excised intron RNA. Their result is shown in Figure 1. The bands in the first lane are size markers. The large bands at the top are the precursor rRNAs. The L and C labels indicate linear and circular forms of the excised intron RNA. Some different conditions of temperature and Mg2+ ion concentration were tried.
The result demonstrated that the splicing reaction was working, but it did not demonstrate RNA catalysis. Why?
The excision reaction depended entirely on the addition of a guanine nucleotide. The researchers explored this requirement further, and some of their results are shown in Figure 2. The rRNA intron excision reaction is shown in the presence of a variety of potential cofactors. The abbreviations for the nucleosides and nucleotides are the standard ones described in Chapter 3. G and dG are the guanosine nucleosides, with dG the 2′-deoxy form.
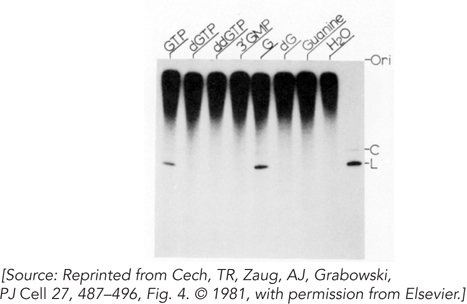
From these results, what can you conclude about the nature of the required cofactor?
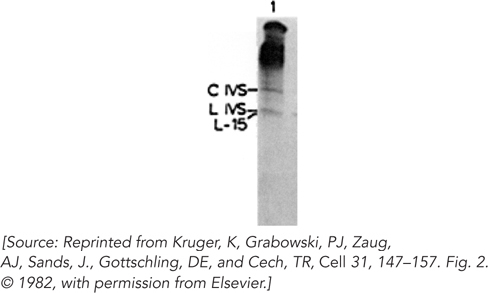
Intrigued that the splicing reaction did not seem to require the addition of cell extract (proteins), the researchers set out to determine whether proteins were required for the reaction. As described in their 1982 paper, they cloned a segment of the gene for the 26S rRNA, including the intron, in a bacterial plasmid and expressed this gene segment in vitro using bacterial RNA polymerase. After deproteinizing the RNA product, they incubated the RNA with the buffer components and guanine nucleotide cofactor, as described in the earlier paper. Their results are shown in Figure 3. The C and L IVS are the circular and linear forms of the excised intron. The L-
Does this experiment demonstrate self-
splicing (RNA catalysis) of the intron? If so, why didn’t the experiment described in the first paper do so?
588