18.3 INITIATION OF PROTEIN SYNTHESIS
Having described the components of the translation machinery, we now turn to a detailed discussion of the initiation stage of protein synthesis—the most highly regulated step of translation. Translation initiation includes recruitment of the small ribosomal subunit to the mRNA; identification of the initiation codon, or start codon; association of the charged initiator tRNA with the mRNA; and recruitment of the large ribosomal subunit to form an active ribosome (Figure 18-15). Each step of protein synthesis, in both E. coli and eukaryotes, requires several protein factors to facilitate the reaction (Table 18-2). Initiation factors (denoted IF in bacteria and eIF in eukaryotes) are critical to enhancing the rate and fidelity of all steps in the process, fine-tuning the underlying rRNA-based activities and the interactions among mRNA, tRNA, and rRNA. Other steps in protein synthesis are generally the same in bacteria and eukaryotes, but some aspects of initiation differ across these two groups.
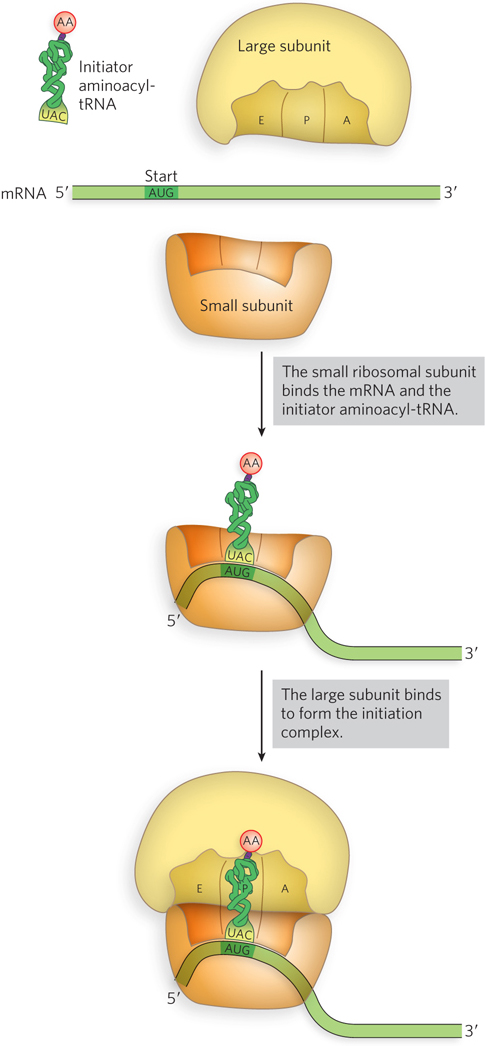
Figure 18-15: An overview of the events in translation initiation.
Base Pairing Recruits the Small Ribosomal Subunit to Bacterial mRNAs
Translation in all organisms begins with binding of the ribosome’s small subunit to an mRNA. In bacteria, the initiating 5′-AUG is guided to its correct position on the ribosome by the Shine-Dalgarno sequence (named for Australian researchers John Shine and Lynn Dalgarno, who identified it), also called the ribosome-binding site (RBS). This consensus sequence is an initiation signal of 4 to 9 purine residues, situated 8 to 13 nucleotides on the 5′ side of the start codon (Figure 18-16a). The sequence base-pairs with a complementary pyrimidine-rich sequence near the 3′ end of the 16S rRNA of the 30S ribosomal subunit. This mRNA-rRNA interaction positions the initiating 5′-AUG sequence of the mRNA in the precise location on the 30S subunit where it is required for translation initiation. Higher degrees of base-pairing complementarity and optimal spacing between the Shine-Dalgarno sequence and the initiation codon increase the efficiency of translation of a particular mRNA. The specific 5′-AUG where the initiator aminoacyl-tRNA—a special type of methionyl-tRNA—is to be bound is distinguished from other methionine AUG codons by its proximity to the Shine-Dalgarno sequence.
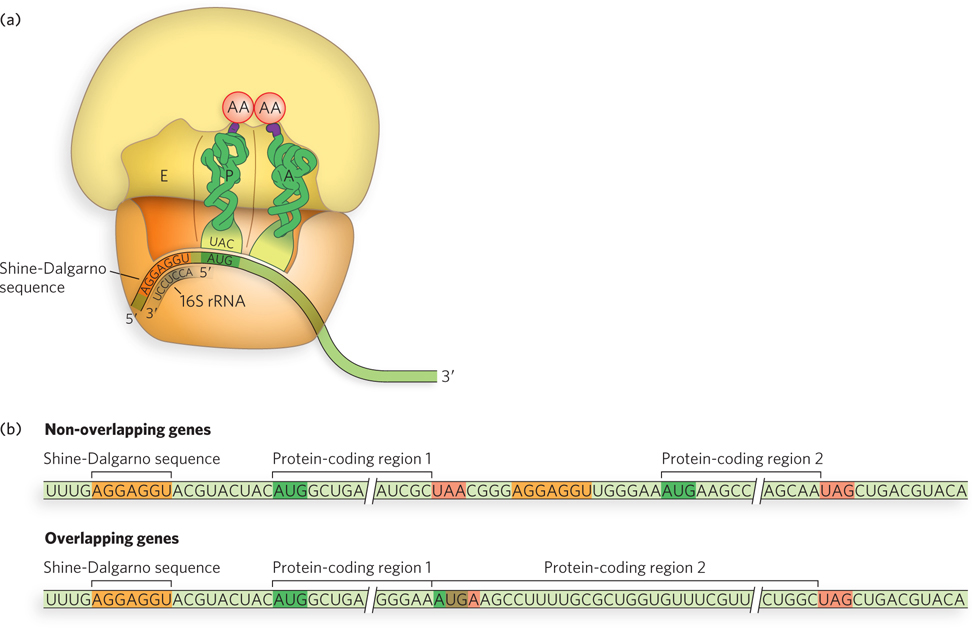
Figure 18-16: The Shine-Dalgarno sequence. (a) The Shine-Dalgarno sequence positions the mRNA on the bacterial ribosome by binding to the 16S rRNA. (b) In bacteria, some polycistronic genes have overlapping start and stop codons (bottom), allowing the ribosome to initiate synthesis in the absence of a Shine-Dalgarno sequence.
The Shine-Dalgarno sequence can be used to initiate synthesis of more than one protein, if they are encoded in a single transcript called a polycistronic mRNA. Most bacterial mRNAs are polycistronic, whereas eukaryotic mRNAs are almost always monocistronic (they encode a single protein). In some bacterial polycistronic mRNAs, the open reading frames overlap (Figure 18-16b, bottom). Despite lacking a Shine-Dalgarno sequence for each internal start site, the internal open reading frames can be translated efficiently because of overlapping start and stop codons, typically 5′-AUGA. Ribosomes terminating translation of the upstream message can initiate the downstream message simply by shifting their reading frame.
Eukaryotic mRNAs Recruit the Small Ribosomal Subunit Indirectly
The 5′ cap and poly(A) tail modifications on eukaryotic mRNAs serve three purposes: they protect the ends from degradation, facilitate nuclear export of the mRNA, and promote translation by binding initiation factors that form a link between the mRNA and the ribosome. The 5′ terminus of the mRNA contains a modified G residue, the 5′ cap, that can be bound by the cap-binding protein, eIF4E, which in turn binds other proteins that recruit the small ribosomal subunit to the mRNA. Once associated with the 5′ end of the mRNA, the small subunit locates the 5′-AUG start codon by scanning the RNA in the 5′→3′ direction.
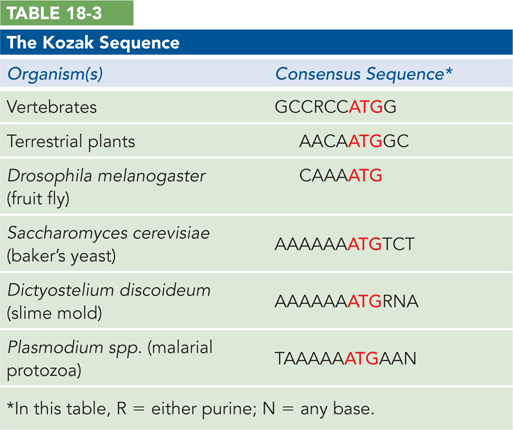
In addition to the 5′ cap, the presence of a purine nucleotide three residues before the start codon and a G residue immediately following the start codon is thought to enhance translation through contact with the initiator tRNA. This Kozak sequence was discovered by Marilyn Kozak during analysis of the sequence features of eukaryotic mRNAs that increased translation efficiency (Table 18-3). At the 3′ terminus of eukaryotic mRNAs, the poly(A) tail stimulates translation efficiency by fostering reinitiation after completion of a polypeptide chain.
A Specific Amino Acid Initiates Protein Synthesis
Protein synthesis begins at the N-terminal end and proceeds by the stepwise addition of amino acids to the C-terminal end of the growing polypeptide, as found by Howard Dintzis in 1961 (see the How We Know section at the end of Chapter 17). The 5′-AUG initiation codon specifies an N-terminal Met residue. Although methionine has only one codon, AUG, all organisms have two tRNAs for methionine. One is used exclusively when 5′-AUG is the initiation codon for protein synthesis; the other is used to code for a Met residue in an internal position in a polypeptide. An initiation factor specifically binds to the initiator tRNA and delivers it to the ribosome, thereby enabling cells to separate translation initiation from elongation.
The distinction between an initiating 5′-AUG and an internal one is straightforward. In bacteria, the two types of tRNA specific for methionine are designated tRNAMet and tRNAfMet. The amino acid incorporated in response to the 5′-AUG initiation codon is N-formylmethionine (fMet). It arrives at the ribosome as N-formylmethionyl-tRNAfMet (fMet-tRNAfMet), which is formed in two successive reactions. First, methionine is attached to tRNAfMet by the Met-tRNA synthetase (which in E. coli aminoacylates both tRNAfMet and tRNAMet), in a reaction of the type shown in Figure 18-11. Second, a transformylase enzyme transfers a formyl group from N10-formyltetrahydrofolate to the amino group of the methionyl part of the tRNA (Figure 18-17). The transformylase is more selective than the synthetase: it is specific for methionyl moieties attached to tRNAfMet, presumably recognizing some unique structural feature of that tRNA. Addition of the N-formyl group to methionine prevents fMet from entering interior positions in a polypeptide (only Met-tRNAMet inserts methionine in interior positions) while allowing fMet-tRNAfMet to bind a specific ribosomal initiation site that accepts neither Met-tRNAMet nor any other aminoacyl-tRNA.
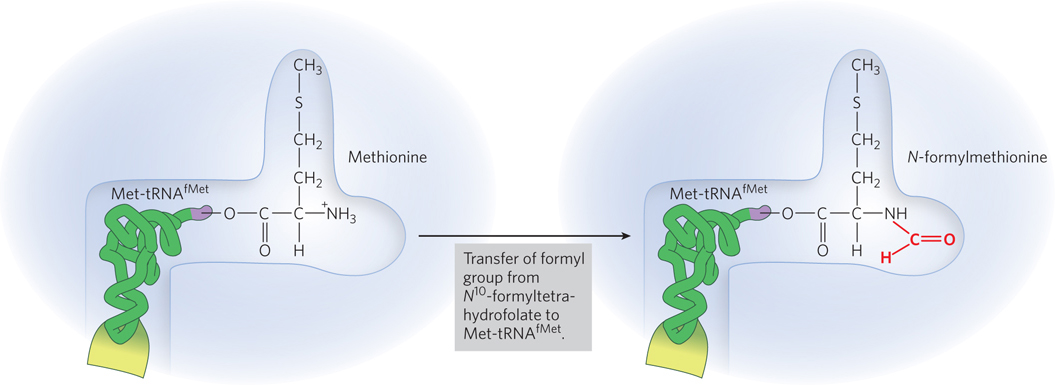
Figure 18-17: Formation of fMet-tRNAfMet. The tRNAfMet is first charged with methionine (not shown), then the Met is converted to fMet by the enzyme methionyl-tRNA formyltransferase (transformylase).
In eukaryotic cells, all polypeptides synthesized by cytosolic ribosomes begin with a Met residue (rather than fMet); however, as in bacteria, the cell uses a special initiator tRNA, tRNAiMet, distinct from the tRNAMet used at AUG codons in interior positions in the mRNA. The Met-tRNAiMet is distinct because it has a specific sequence in the anticodon arm that is recognized by an initiation factor protein. Polypeptides synthesized by mitochondrial and chloroplast ribosomes, however, begin with N-formylmethionine. This strongly supports the view that mitochondria and chloroplasts originated from bacterial ancestors symbiotically incorporated into precursor eukaryotic cells at an early stage of evolution.
In bacteria, the enzyme deformylase typically removes the formyl group from the N-terminal Met residue during or shortly after production of the polypeptide. In both bacteria and eukaryotes, enzymes called aminopeptidases frequently remove the entire N-terminal methionine, and sometimes one or two additional amino acids, from newly synthesized polypeptides. Thus, many mature proteins do not have a Met residue at their N-terminal end.
Initiation in Bacterial Cells Requires Three Initiation Factors
As discussed above, ribosomes have three sites that bind aminoacyl-tRNAs: the A, P, and E sites. In addition to the 30S subunit, an mRNA, and an initiating fMet-tRNAfMet, the initiation of polypeptide synthesis in bacteria requires a set of three initiation factor proteins known as IF-1, IF-2, and IF-3. Each plays a specific role in assembling the small ribosomal subunit (with the mRNA and initiator tRNA in place) and the large ribosomal subunit in a process controlled by GTP hydrolysis.
Formation of an active 70S ribosome takes place in three steps, as shown in Figure 18-18. In step 1, the 30S subunit binds two initiation factors, IF-1 and IF-3. IF-3 prevents premature combination of the 30S and 50S subunits. IF-1 binds at the A site and blocks tRNA binding during initiation. The mRNA then binds to the 30S subunit through base pairing of the Shine-Dalgarno sequence with 16S rRNA. This short mRNA-rRNA helix is bound in a cleft in the 30S subunit, which precisely positions the mRNA adjacent to the P site, thus accounting for the accuracy of start codon selection. The initiating 5′-AUG is now positioned at the P site. This is the only site to which fMet-tRNAfMet can bind, and fMet-tRNAfMet is the only aminoacyl-tRNA that can bind first to the P site. During the subsequent elongation stage, all other incoming aminoacyl-tRNAs (including the so-called elongator Met-tRNAMet that binds interior AUG codons) bind first to the A site and only subsequently transfer to the P and E sites.
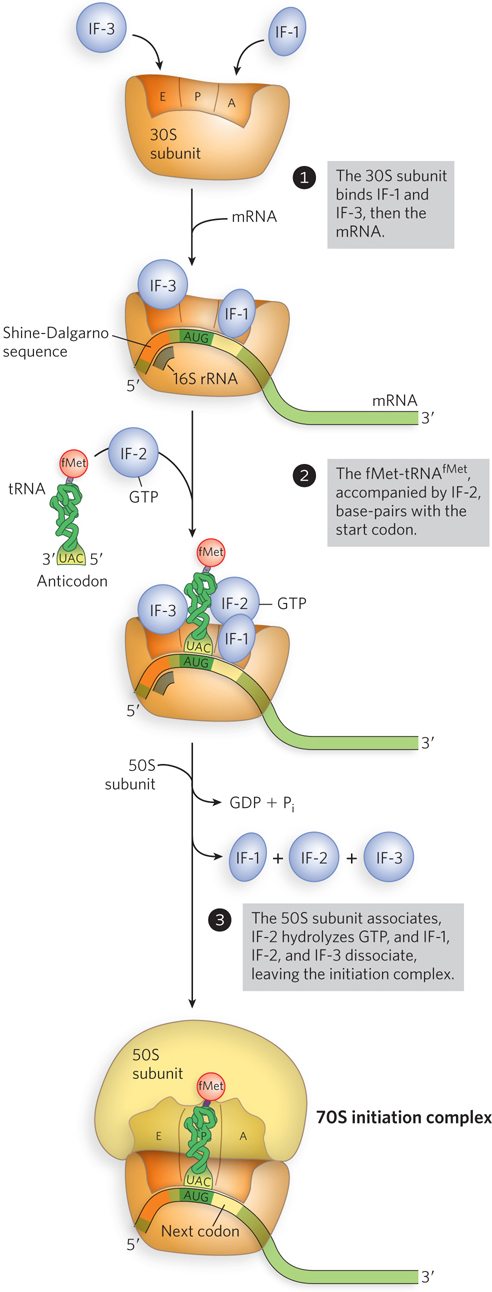
Figure 18-18: Translation initiation in bacteria. Initiation occurs in three steps. Initiation factors IF-1 and IF-3 prevent premature binding of the elongation aminoacyl-tRNAs (i.e., those used at the elongation stage) and the 50S subunit, respectively. IF-2 accompanies fMet-tRNAfMet to the initiation site.
In step 2, the complex consisting of the 30S ribosomal subunit, IF-1, IF-3, and mRNA is joined by both GTP-bound IF-2 and the initiating fMet-tRNAfMet. The anticodon of this tRNA now pairs with the mRNA’s start codon in the P site. X-ray crystallography has revealed contacts between rRNA bases and three G≡C base pairs in the anticodon arm of initiator, but not elongator, Met-tRNAs. This observation suggests a mechanism by which initiator tRNA is favored in the P site during the initiation process. In step 3, a conformational change in the 30S subunit triggers the release of IF-3, enabling association with the 50S subunit; simultaneously, the GTP bound to IF-2 is hydrolyzed to GDP and Pi, which are released from the complex. All three initiation factors depart from the ribosome at this point.
Completion of the steps in Figure 18-18 produces a functional 70S ribosome called the initiation complex, containing the mRNA and the initiating fMet-tRNAfMet. The correct location of fMet-tRNAfMet in the P site in the initiation complex is ensured by at least three points of recognition and attachment: (1) the codon-anticodon interaction involving the initiation 5′-AUG fixed in the small subunit portion of the P site, (2) interaction between the Shine-Dalgarno sequence in the mRNA and the 16S rRNA of the small subunit, and (3) binding interactions between the large-subunit portion of the P site and the fMet-tRNAfMet. The initiation complex is now ready for elongation.
Initiation in Eukaryotic Cells Requires Additional Initiation Factors
Translation is generally similar in eukaryotic and bacterial cells; most of the significant differences are in the mechanism of initiation. Eukaryotic initiation requires, besides separate small and large ribosomal subunits, at least 12 initiation factors and the binding and hydrolysis of ATP and GTP, as illustrated in Figure 18-19. In step 1, before translation begins, the ribosomal subunits are separated by initiation factors eIF3 and eIF1A; eIF1A is the functional homolog of IF-1 in bacteria. These eukaryotic factors prevent premature subunit joining and block initiator tRNA binding to the ribosomal A site, respectively. A third initiation factor, eIF1, binds to the E site. Meanwhile (step 2), the GTP-binding factor eIF2—containing three subunits, eIF2α, eIF2β, and eIF2γ—associates with GTP and a charged initiator tRNA (Met-tRNAiMet) to form an eIF2–GTP–Met-tRNAiMet ternary complex. An A=U base pair near the amino acid–binding site of the amino acid arm of Met-tRNAiMet, but not present in elongator Met-tRNAMet, is critical for binding to eIF2-GTP. Two other proteins, eIF5 (not shown in Figure 18-19) and eIF5B, which are involved in later steps of ribosomal assembly, also associate at this point. Three factors, eIF1, eIF1A and eIF3, mediate interaction between the ternary complex and the 40S subunit to form a 43S preinitiation complex.
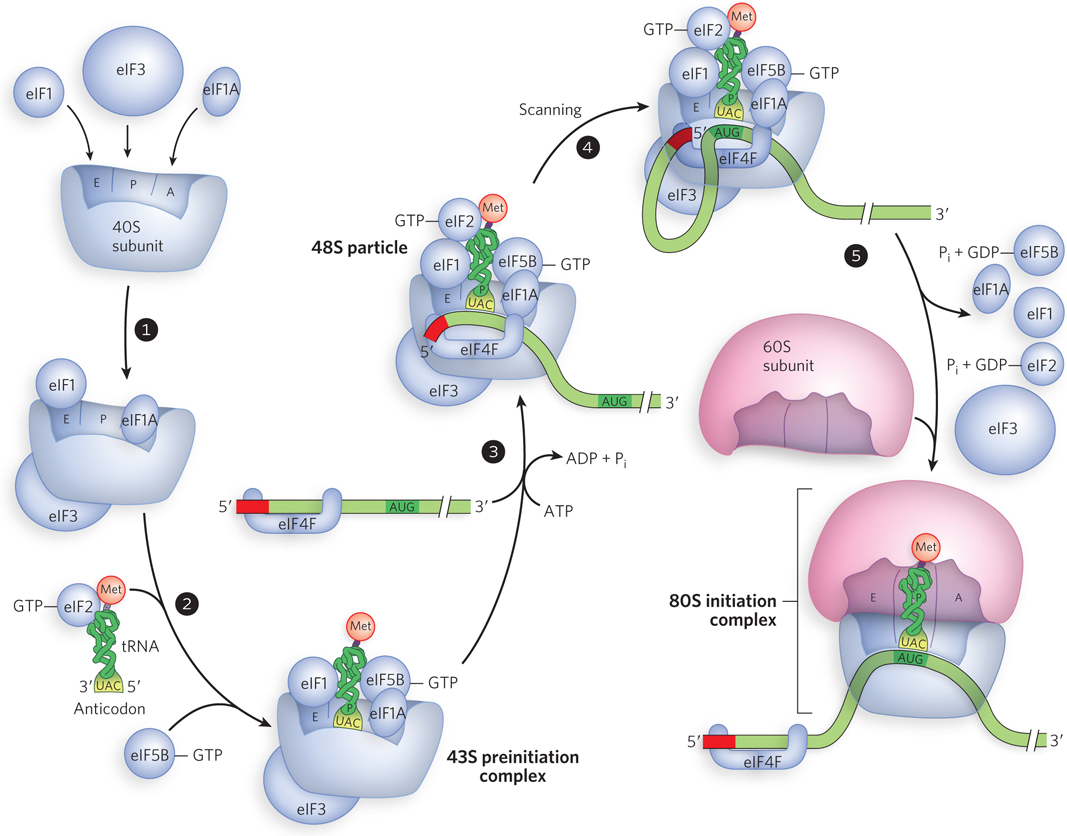
Figure 18-19: Translation initiation in eukaryotes. Initiation is more complex in eukaryotes. Eukaryotic initiation factors (eIFs) promote the assembly of the 43S preinitiation complex with an mRNA and subsequent scanning of the mRNA to identify the start codon. Note that circularization of the mRNA in step 3 is omitted here for clarity. It is shown in Figure 18-20.
Binding of the 43S preinitiation complex to an mRNA (step 3) is mediated by a complex called eIF4F. It contains eIF4E, a factor that binds the 5′ cap; eIF4A, an ATPase and RNA helicase; and eIF4G, which binds both eIF4E and eIF3 to provide a link between the 43S complex and the mRNA. The eIF4G also binds to poly(A) binding protein (PABP), which is associated with the 3′ poly(A) tail of the mRNA, and this eIF4G-PABP association brings the 5′ and 3′ ends of the mRNA together (Figure 18-20). Circularization of the eukaryotic mRNA by the eIF4G-PABP interaction also facilitates the translational regulation of gene expression (as described in Chapter 21). Following association of the eIF4F complex, another initiation factor, eIF4B, binds (not shown in Figure 18-19); its function is less clear.
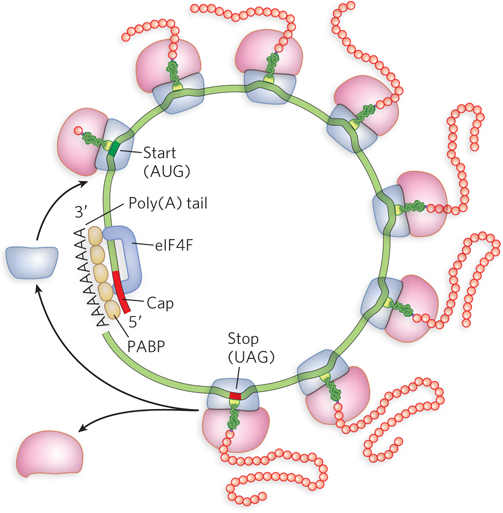
Figure 18-20: Circularization of eukaryotic mRNA. eIF4F (purple) consists of eIF4A, eIF4E, and eIF4G. In step 3 of the initiation process (see Figure 18-19), eIF4G binds to the 5′ cap and the poly(A) binding protein (PABP), effectively circularizing the mRNA.
This larger complex, containing the 43S components, eIF4F, and mRNA, is a stable 48S particle. Within the 48S particle, the 43S complex is capable of scanning, sliding along the mRNA in search of a start codon. Once assembled on the 5′ end of the mRNA (step 4 in Figure 18-19), the 43S complex scans along the mRNA in the 5′→3′ direction to the first AUG, which is almost always recognized as the start site for translation initiation. The eIF4F complex is probably involved in this scanning process, perhaps using the RNA helicase activity of eIF4A to eliminate secondary structure in the 5′ untranslated portion of the mRNA. Scanning is also facilitated by eIF4B, but the details of this process are unknown. Base pairing between the start codon and the anticodon of the initiator tRNA is primarily responsible for identification of the initiation site. Factor eIF1 is also critical to proper AUG recognition by preventing stable ribosomal association with non-AUG codons.
When the 43S complex has located the AUG start codon in a bound mRNA, the complex joins the 60S subunit to form an active 80S ribosome (step 5 in Figure 18-19). Because eIF1, eIF1A, and eIF2 occlude parts of the 40S subunit surface that must interface with the 60S subunit, they must be displaced before subunit joining. This process requires eIF5 and eIF5B. The GTPase-activating eIF5 is specific for eIF2, stimulating hydrolysis of the eIF2-bound GTP and thus reducing eIF2’s affinity for the initiator tRNA. Finally, eIF5B, a ribosome-dependent GTPase homologous to bacterial IF-2, hydrolyzes GTP and triggers dissociation of eIF2-GDP and other initiation factors from the 40S subunit, with concomitant association of the 60S subunit to form the functional 80S ribosome.
The initiation complex is now complete. The efficiency of translation is affected by many properties of the mRNA and proteins in this complex, including the length of the 3′ poly(A) tail (in most cases, longer is better).
Some mRNAs Use 5′ End–Independent Mechanisms of Initiation
Some viral and eukaryotic mRNAs lack a 5′ cap but still rely on the eukaryotic translation machinery to produce their proteins. They accomplish this with an RNA segment called an internal ribosome entry site (IRES), located on the 5′ side of the start codon; it recruits the 40S subunit through direct interaction with the subunit or with eIF4F (Figure 18-21a). Cap-independent binding can also involve other proteins, such as La protein, which binds a pyrimidine-rich segment in the 5′ region of the mRNA.
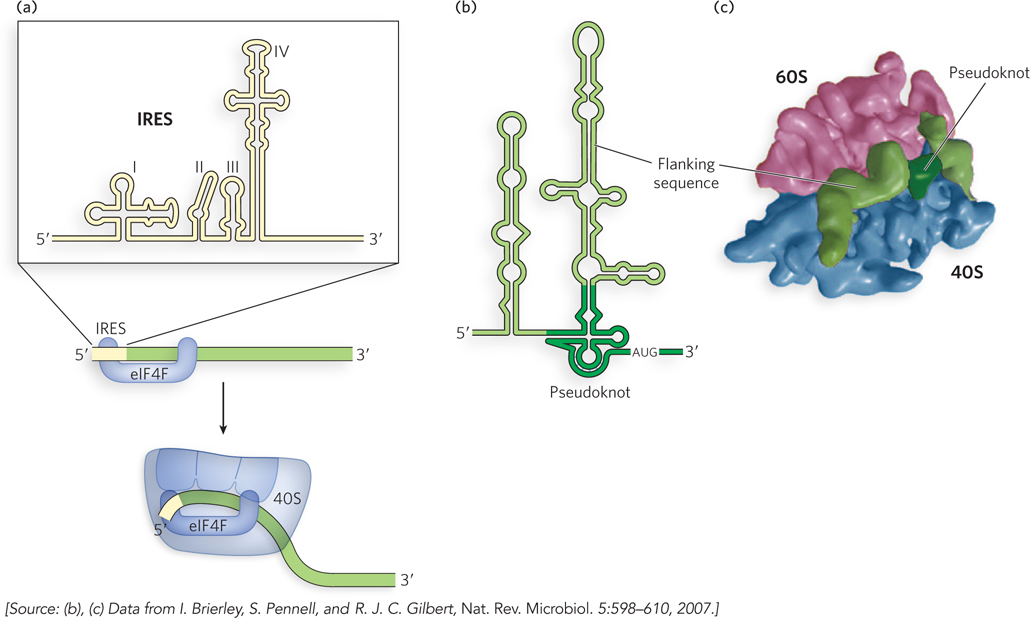
Figure 18-21: Internal ribosome entry sites of viruses. (a) In poliovirus mRNA, the IRES is brought to the translation machinery by the host cell’s eIF4F. (b) The IRES of hepatitis C virus contains a pseudoknot structure. (c) The human 80S ribosome (pink and blue) binds to the HCV IRES through its pseudoknot and flanking sequences (green).
The first IRES was discovered in poliovirus, when researchers noticed that the viral mRNA is efficiently translated despite lacking a 5′ cap. On infection, poliovirus produces a protease that cleaves the host cell’s eIF4G into two fragments. This renders eIF4G useless for the host cell’s protein synthesis but does not compromise viral protein synthesis, because the poliovirus IRES requires just one fragment of eIF4G to initiate translation. In this way, the virus can simultaneously reduce levels of host protein expression and maintain its own protein synthesis in infected cells. Different viral IRES subtypes seem to form distinct three-dimensional structures that enable direct interactions with the host translation machinery. For example, hepatitis C virus (HCV) mRNA has a pseudoknot adjacent to two stem-loop structures (Figure 18-21b). The resulting three-dimensional structure binds to the host cell’s 80S ribosome such that the start codon is positioned in the P site (Figure 18-21c).
Some cellular mRNAs, although they contain a 5′ cap, also contain IRES segments that enable translation under conditions that normally block initiation. Cellular mRNAs bearing an IRES can be translated even during viral infection and in other situations in which most protein synthesis is shut off, such as during starvation and programmed cell death. This is an important advantage of the IRES-mediated initiation mechanism. Internal initiation avoids the use of the 5′ cap and the initiation factors needed to recognize it. The IRES positions the mRNA start codon correctly on the 40S subunit to ensure accurate initiation during each round of translation.
SECTION 18.3 SUMMARY
In bacteria, ribosomes are recruited to the mRNA by the Shine-Dalgarno sequence, 4 to 9 purine residues located 8 to 13 nucleotides on the 5′ side of the start codon.
In eukaryotes, the mRNA 5′ cap is bound by initiation factor eIF4E, which then binds other proteins that recruit the ribosomal subunits. The ribosome identifies the initiation codon by scanning the mRNA in the 5′→3′ direction.
In bacteria, the initiating aminoacyl-tRNA in all proteins is N-formylmethionyl-tRNAfMet. In eukaryotic cells, it is a special form of methionyl-tRNA, Met-tRNAiMet.
Bacterial initiation factors IF-1, IF-2, and IF-3 promote assembly of the mRNA, fMet-tRNAfMet, and both ribosomal subunits to form the initiation complex. IF-1 and IF-3 prevent premature binding of the large subunit and elongator tRNAs, respectively. IF-2, a GTPase, recruits fMet-tRNAfMet to the P site. GTP hydrolysis allows the release of all three initiation factors and promotes association of the large subunit.
Initiation in eukaryotes involves a host of initiation factors: eIF1, eIF1A, and eIF3 promote association of the small ribosomal subunit with eIF2-bound Met-tRNAiMet, eIF5, and eIF5B, forming the 43S preinitiation complex. The mRNA 5′ cap is bound by the eIF4F complex, which also binds to the small ribosomal subunit. Once this 48S particle is formed, the small subunit scans the mRNA for the start codon, after which the large subunit associates to form an active 80S ribosome. Circularization of the eukaryotic mRNA involves interactions between factors bound at the 5′ end (eIF4G) and the 3′ end (PABP).
Some viral and eukaryotic mRNAs do not depend on the 5′ cap for translation initiation, but instead bind eukaryotic initiation factors and the 40S ribosomal subunit at an internal ribosome entry site (IRES) downstream from the 5′ end.