19.1 REGULATION OF TRANSCRIPTION INITIATION
Regulatory processes operating at the level of transcription initiation are the best documented, and probably the most common. Elaborate mechanisms have evolved to regulate the process of transcription initiation—
Regulatory proteins that bind DNA can have profound effects on the affinity of RNA polymerase for a promoter. These effects can flow in either direction, either enhancing or preventing RNA polymerase function. Given both the energy expenditure of gene expression and the need for cells to be able to respond quickly to changes in their environment, one can only imagine the enormous evolutionary pressure placed on regulatory mechanisms. We provide here an overview of the transcriptional regulatory mechanisms used by cells. The most detailed information derives from studies in bacterial systems, but eukaryotic mechanisms of gene regulation, although more complex and using somewhat different strategies, can be explained by the same basic principles.
Activators and Repressors Control RNA Polymerase Function at a Promoter
The most basic mechanism for regulation of transcription initiation is encoded in the DNA sequence of the promoter. RNA polymerase has different intrinsic affinities for promoters of different sequence. In the absence of other controls, these differences in promoter strength correlate with the efficiency with which the genes are transcribed. Genes for products that are required at all times, such as the enzymes of central metabolic pathways, are expressed at a nearly constant level. These genes are often referred to as housekeeping genes, and unvarying expression is called constitutive gene expression. Although housekeeping genes are expressed constitutively, the expression levels of different housekeeping genes vary widely. For these genes, the RNA polymerase–
When the level of a gene product rises and falls with a cell’s changing needs, this is known as regulated gene expression. Activation is an increase in expression and repression is a decrease in expression of a gene in response to a change in environmental conditions. The mechanisms of gene activation and repression, in both bacteria and eukaryotes, require the assistance of transcription factors (also called transcription regulators), proteins that alter the affinity of the RNA polymerase for the promoter. Transcription factors that enhance gene expression are called activators, and those that reduce expression are called repressors. As we will see later in this chapter, bacterial and eukaryotic transcription factors have many common structural and functional features. These regulators act by binding to specific DNA sequences known as regulatory sites.
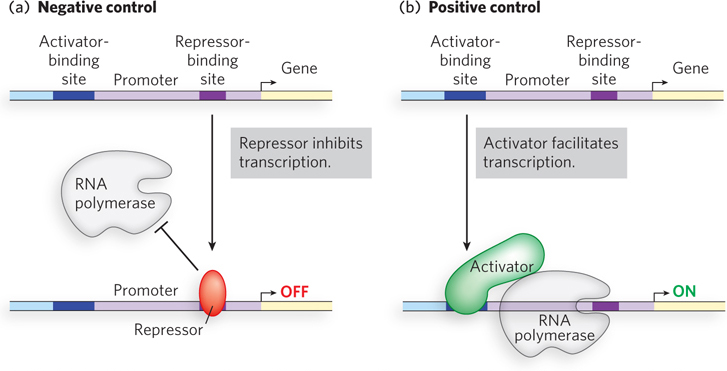
A gene is said to be under positive regulation when binding of an activator protein promotes or increases expression of that gene. Conversely, a gene is under negative regulation when binding of a repressor protein prevents or decreases expression. Thus, positive and negative regulation refer to the type of regulatory protein involved: the bound protein either facilitates or inhibits transcription.
A repressor can lower the rate of gene transcription if its regulatory site overlaps the gene’s promoter and repressor binding sterically occludes binding of the RNA polymerase to the promoter (Figure 19-2a). Repressors can act in other ways as well. Some prevent transcription by binding a regulatory site that is near the promoter, but the binding itself does not block RNA polymerase binding. Recall that the RNA polymerase–
668
Activators provide a molecular counterpoint to repressors: they bind to DNA and enhance the activity of RNA polymerase at a promoter. For example, an activator may induce a conformational change in the polymerase that accelerates transition to the open complex. Alternatively, an activator may alter the torsion of DNA, making it more likely to unwind and form the open complex. Probably the most common way that activators function is through cooperativity (Figure 19-2b). In this case, the activator binds both the RNA polymerase and a DNA regulatory site next to the promoter, thereby increasing the affinity of the polymerase for the promoter; this activation process is referred to as recruitment.
Transcription Factors Can Function by DNA Looping
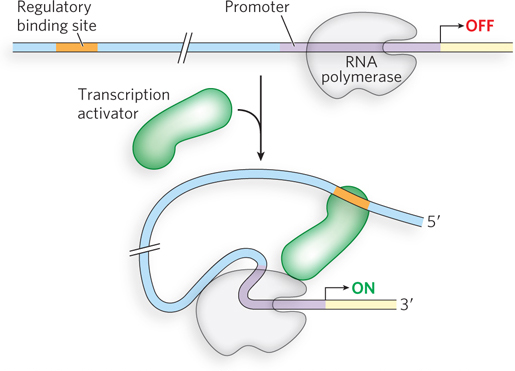
Binding sites for activators and repressors are often found at or near the promoter, particularly in bacteria. However, regulatory sites can also be found far away from the promoter. In fact, in eukaryotes, regulatory sites are sometimes thousands of base pairs upstream or downstream from a promoter. How do transcription factors exert their effects on RNA polymerase when their binding sites are so far away from the gene’s promoter? Experiments directed at understanding this “action at a distance” have demonstrated that distant regulatory sites can often be placed closer to or farther from the promoter and still retain their function. Not only is distance from the promoter of little consequence (provided it is not too close and overlaps the promoter), but the regulatory sites also retain their function regardless of experimental changes in their sequence orientation relative to the promoter.
When distant regulatory sites were first discovered, scientists imagined that the regulatory proteins might bind to these sites and then slide along the DNA until they reached RNA polymerase at the promoter. However, experiments revealed that this was not the case (see the How We Know section at the end of this chapter). Instead, the DNA between the regulatory site and the RNA polymerase–
669
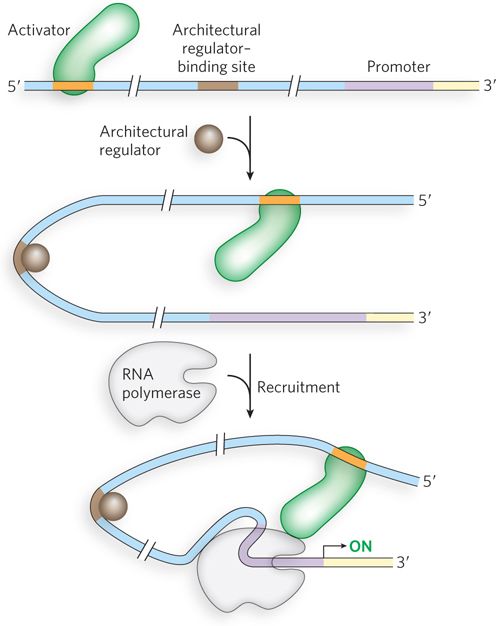
Gene regulation by DNA looping can result in either activation or repression. Activation can recruit RNA polymerase to the promoter through cooperativity, in much the same way as an activator that binds near the promoter. Recruitment of RNA polymerase through DNA looping can also be mediated by a protein “bridge” between the activator and the polymerase (Figure 19-6a). Proteins that act by bridging activators and RNA polymerase, but do not bind DNA directly, are called coactivators. For example, the eukaryotic protein complex Mediator acts as a bridge between RNA polymerase II and regulatory proteins bound to distant sites and is essential for transcription activation (see Chapter 15). Repression can also occur through proteins that do not bind the DNA directly but instead bind activator proteins and prevent the recruitment of RNA polymerase (Figure 19-6b). Repressors that act through protein-
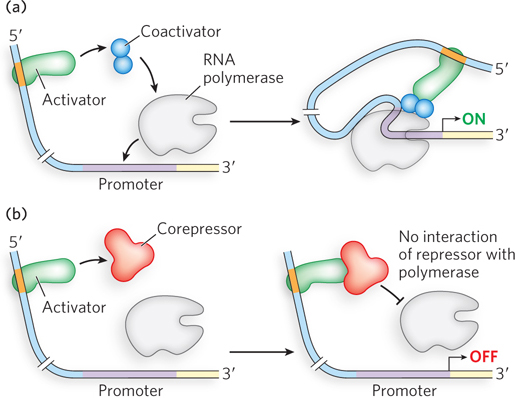
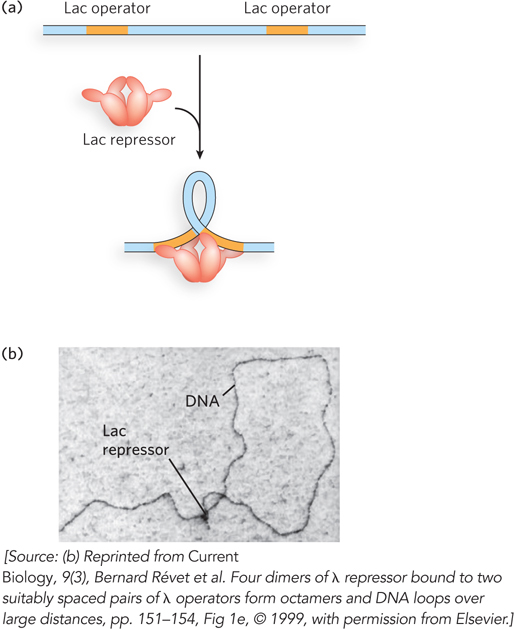
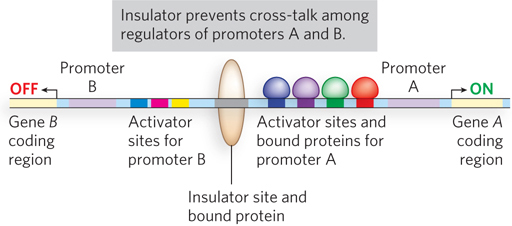
There could be an unintended consequence of gene regulation that uses DNA loops over large distances. A regulator meant to target a distant promoter could act instead on a different promoter located in the opposite direction. In eukaryotes, this problem is solved by the presence of insulators, short sequences of DNA that prevent inappropriate cross-
670
Regulators Often Work Together for Signal Integration
Activators and repressors often function at the same promoter. The use of multiple transcription factors allows the expression of a gene to be affected by more than one environmental condition. Signal integration, occurring in both eukaryotes and bacteria, is the control of a gene by multiple regulators in response to more than one environmental signal. A simple example in bacteria is the regulation of genes that encode products responsible for metabolizing sugar, the main energy source for bacteria (Figure 19-8).
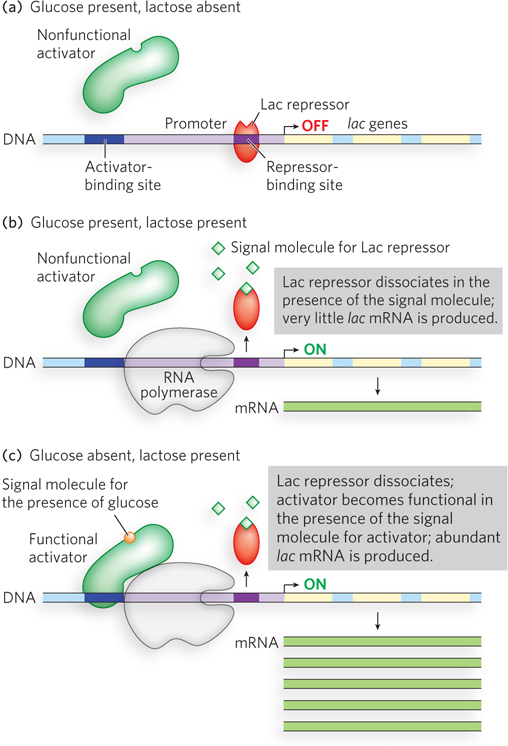
Bacteria can derive energy from many different sugars, and they have sets of genes for metabolizing each one. But it would be a waste of cellular resources to express all of these genes all the time, and systems of regulation have evolved in which the genes for metabolizing a given sugar are expressed only when that sugar is present in the environment. Take, for example, the lac operon, a set of genes for the metabolism of lactose (see Chapter 5 and Chapter 15; operons are more fully defined later in this section). When lactose is not present, the Lac repressor protein is bound to the operon DNA at a sequence called the operator and ensures that the genes for lactose metabolism are not transcribed. When lactose is present, the cell sends a signal for the Lac repressor to dissociate from the operator, allowing transcription of the genes encoding lactose-
671
Though bacteria can metabolize many different sugars, their best energy source is glucose. When both glucose and lactose are present in the environment, the cell preferentially metabolizes glucose. It would be a waste of energy to continue producing the lactose-
This exquisite control, achieved by two different transcription factors working together, is an example of signal integration. The cell can adjust its energy resources by taking into account more than one environmental condition (the availability of glucose and of lactose).
Gene Expression Is Regulated through Feedback Loops
The regulation of gene expression usually operates as a feedback circuit. This is easier to explain in bacteria than in eukaryotes, although similar principles apply in both. Recall that genes for the metabolism of lactose are controlled by multiple transcription factors. The repressors and activators either bind DNA or do not, depending on signals received from the environment. The binding of a repressor or activator to DNA is often regulated by a molecular signal called an effector, usually a small molecule or another protein that binds the activator or repressor and causes a conformational change that results in an increase or decrease in transcription.
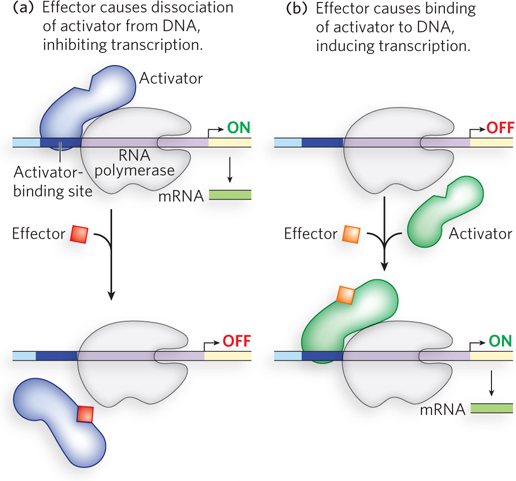
Repressors can be activated or inactivated by effectors. In one scenario, the effector binds to the repressor and induces a conformational change that results in dissociation of the repressor from its binding site on the DNA, allowing transcription to proceed (Figure 19-9a). Alternatively, the interaction of an inactive repressor and a signal molecule could cause the repressor to bind to DNA, shutting down transcription (Figure 19-9b).
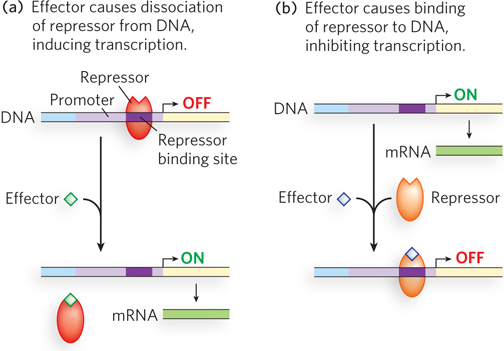
The same considerations apply to activators. Some activators bind DNA and enhance transcription until dissociation of the activator is triggered by the binding of a signal molecule (Figure 19-10a). In other cases, the activator binds to DNA only after interaction with a signal molecule (Figure 19-10b). Signal molecules that bind activators can therefore increase or decrease transcription, depending on how they affect the activator.
672
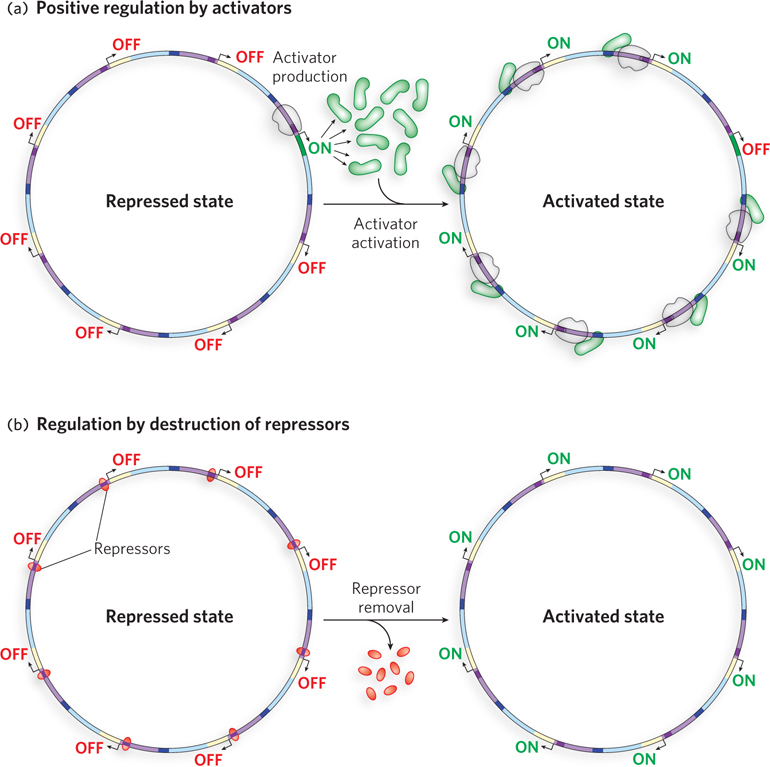
Given the allosteric control of activators and repressors, we can understand how a regulatory feedback loop functions. In the bacterial lac operon, Lac repressor binds DNA in the absence of an effector, preventing the expression of genes required for the metabolism of lactose. The effector for the Lac repressor is allolactose, a minor byproduct of lactose metabolism. Therefore, when lactose is present in the environment, the signal molecule is formed and binds the Lac repressor, causing it to dissociate from the DNA. This gives RNA polymerase access to the promoter of the lac operon for a low, basal level of transcription. Transcription of the operon is greatly enhanced by binding of the activator cAMP receptor protein (CRP). CRP does not bind its regulatory site when glucose is available. In the absence of glucose, however, cells produce cAMP (cyclic AMP), which is an allosteric effector of CRP, producing a conformational change that enables CRP to bind its regulatory site. The bound activator then recruits RNA polymerase and boosts gene expression from the lac operon. A second level of control, called inducer exclusion, occurs when the lactose transporter is blocked by the glucose permease. The glucose permease is usually phosphorylated, and it becomes dephosphorylated on the transfer of phosphate to glucose during glucose transport. The dephosphorylated form of the permease binds and directly blocks the lactose and maltose transporters. When glucose is absent, the glucose permease exists mainly in its phosphorylated form and no longer inhibits the transporters of other sugars.
When lactose is depleted, allolactose is also depleted, and in the absence of this effector the Lac repressor again binds the operator site, preventing RNA polymerase from transcribing the lac operon. Likewise, when glucose becomes available, cAMP levels diminish and CRP no longer binds DNA. Regulatory feedback loops like these are common in all cells.
Related Sets of Genes Are Often Regulated Together
Bacterial promoters are often positioned upstream from several genes that operate in a common metabolic pathway. Transcription produces a long polycistronic mRNA that contains multiple genes in one transcript. The single promoter that initiates transcription of the cluster is the site of regulation for all the genes in the polycistronic message. The polycistronic DNA, its promoter, and all the additional sequences that function together in regulating its transcription are called an operon (Figure 19-11). Most operons contain 2 to 6 genes, but some have more than 20 genes.
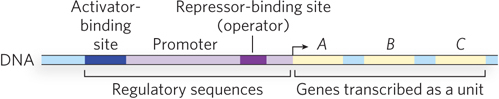
The organization of bacterial genes into operons allows small sets of genes that function together to be regulated together. But there are also instances in which multiple operons are controlled in a coordinated fashion. A group of operons with a common regulator is called a regulon. This arrangement allows shifts in cellular functions that can require the action of hundreds of genes—
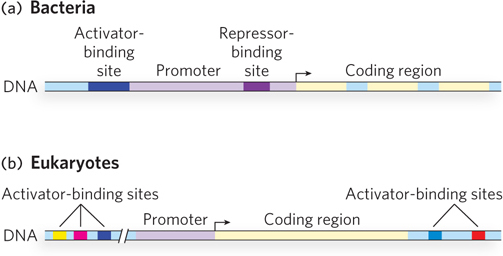
Eukaryotic Promoters Use More Regulators Than Bacterial Promoters
Signal integration is important to gene regulation in both bacteria and eukaryotes. However, eukaryotic promoters for Pol II, the RNA polymerase that transcribes protein-
673
Multiple Regulators Provide Combinatorial Control
Using multiple transcription factors for every gene in a genome would be energetically costly if every gene required unique regulators, but the use of different combinations of a limited set of transcription factors to differentially regulate many genes provides an opportunity for efficiency. This is made possible by combinatorial control—the need for specific combinations of factors to unlock each particular gene (Figure 19-14). Consider the hypothetical genes A, B, and C, each of which requires five transcription factors. If each factor were distinct, the cell would require 15 transcription factors to control the expression of these three genes. But if genes A and B used three of the same factors, and a combination of the factors for genes A and B is used to regulate gene C, then differential regulation of these three genes would require only 7 different proteins instead of 15.
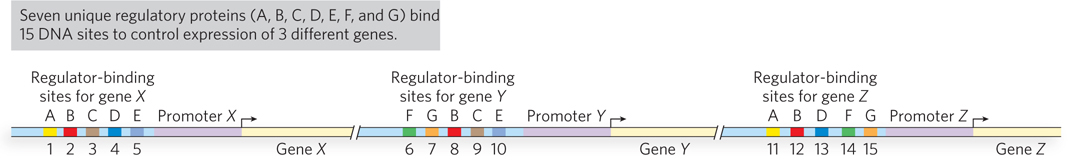
674
Combinatorial control occurs in bacteria as well as in eukaryotes, and we have already seen an example in bacteria in the case of the two regulatory elements of the genes involved in lactose metabolism. Recall that these genes are controlled by a repressor that senses the presence of lactose and by an activator that senses the presence of glucose. The genes encoding proteins for the metabolism of other sugars have their own repressors, but they use the same activator. For instance, the digestion of galactose requires removal of the galactose repressor from the DNA, and this occurs only when galactose is present. However, as with the lactose genes, high expression of the galactose genes is achieved only when glucose is absent from the environment. The same protein activator used at the lac operon also regulates the galactose genes: CRP, which becomes functional by binding its effector molecule cAMP when glucose is not present. The regulation of the genes for different sugar-
Regulation by Nucleosomes Is Specific to Eukaryotes
In eukaryotes, transcription initiation almost always depends on the action of activator proteins. One important reason for the apparent predominance of positive regulation seems obvious: packaging of DNA into chromatin renders most promoters inaccessible, and thus their associated genes are silent. Chromatin structure affects access to some promoters more than others, but generally, repressors that prevent the access of RNA polymerase to DNA would be redundant. Therefore, eukaryotic genes are constitutively repressed and require activation in order to be transcribed. Recall from Chapter 10 that transcription is regulated by different types of change in chromatin structure. The chromatin state can be either open or closed. Open chromatin is often (but not always) associated with acetylation of nucleosomes, whereas closed chromatin is associated with methylation of nucleosomes. Thus eukaryotic activators and repressors can act through modification of nucleosomes that alter chromatin structure, rather than by recruiting RNA polymerase or preventing polymerase binding to DNA.
Bacterial RNA polymerase generally has access to every promoter, and most bacterial genes are controlled by specific repressors. In eukaryotes, however, general repression by nucleosomes, combined with the use of activators to regulate transcription, is more efficient than the use of specific repressors. If the 20,000 to 25,000 genes in the human genome were negatively regulated, each cell would have to constantly synthesize specific repressors to prevent the transcription of a great many genes. Instead, the nucleosomes that function to condense DNA also repress most genes, and the cell has to synthesize only the activators needed to promote transcription of the subset of genes required at a particular time. These arguments notwithstanding, there are examples of negative regulation in eukaryotes, from yeast to humans.
SECTION 19.1 SUMMARY
The various mechanisms of transcription initiation are among the most well-
documented regulated processes in gene expression. Transcription initiation is the step most often regulated, and regulation at this point is the most energy efficient, because it occurs before the investment of energy in mRNA and protein synthesis. 675
Transcription initiation is mediated by intrinsic promoter affinity for RNA polymerase or by repressor and activator proteins that modulate promoter affinity for the polymerase.
Repressors can hinder transcription by binding DNA at a site that prevents RNA polymerase binding or by preventing the closed-
to- open transition of the polymerase- promoter complex (negative regulation). Activators promote RNA polymerase binding through cooperativity or promote formation of the open complex by causing a conformational change in the promoter or the polymerase (positive regulation).
Binding sites for transcription factors need not be close to the transcription start site, and in eukaryotes they are often located thousands of base pairs from the promoter. Regulatory proteins that bind sites distant from the promoter exert their effects through DNA looping.
Promoters may be controlled by two or more transcription factors, allowing integration of signals from more than one environmental variable.
Small signal molecules (effectors) allosterically regulate the function of activators and repressors.
Sets of genes that function in one pathway are often controlled simultaneously.
Eukaryotes generally have more transcription factors than bacteria, reflecting the greater need for regulated gene expression in a complex multicellular organism. Specificity of gene expression is enhanced by the use of multiple regulators.
In combinatorial control, the same regulatory protein is used to control different genes in combination with other regulators, forming a multiprotein regulator that is specific for individual genes.
Chromatin structure renders most eukaryotic promoters inaccessible to RNA polymerase and plays an important role in gene expression, which typically requires proteins that modify nucleosomes and open up the chromatin structure to transcriptional activation.