3.4 FATE OF THE RNA PRIMARY TRANSCRIPT
The RNA transcript that comes off the template DNA strand is known as the primary transcript, and it contains the genetic information of the gene that was transcribed. For protein-coding genes, this means that the primary transcript includes the information needed to direct the ribosome to produce the protein corresponding to the gene (Chapter 4). The RNA molecule that combines with the ribosome to direct protein synthesis is known as the messenger RNA (mRNA) because it serves to carry the genetic “message” (information) from the DNA to the ribosome. As we will see in this section, there is a major difference between prokaryotes and eukaryotes in the manner in which the primary transcript relates to the mRNA. We will also see that some genes do not code for proteins, but for RNA molecules that have functions of their own.
3.4.1 Messenger RNA carries information for the synthesis of a specific protein.
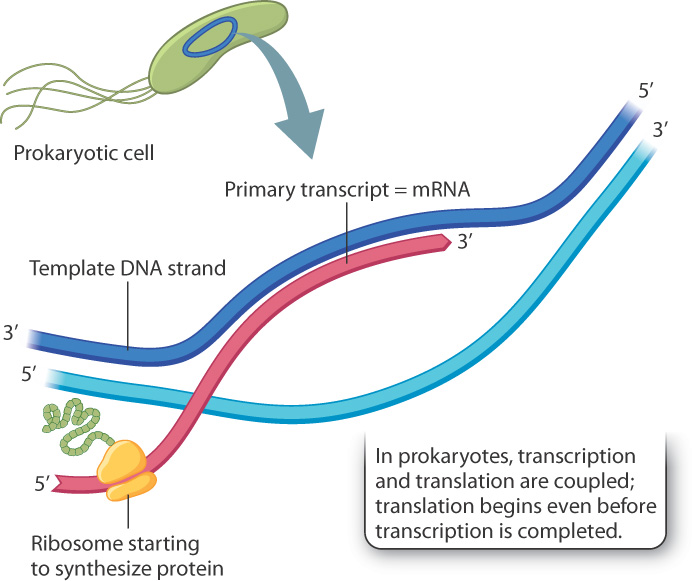
In prokaryotes, the relation between the primary transcript and the mRNA is as simple as can be: The primary transcript is the mRNA. Even as the 3′ end of the primary transcript is still being synthesized, ribosomes bind with special sequences near its 5′ end and begin the process of protein synthesis (Fig. 3.21). This intimate connection between transcription and translation can take place because prokaryotes have no nuclear envelope to spatially separate transcription from translation; the two processes are coupled, which means that they are connected in space and time.
3-16
Primary transcripts for protein-coding genes in prokaryotes have another feature not shared with those in eukaryotes: They often contain the genetic information for the synthesis of two or more different proteins, usually proteins that code for successive steps in the biochemical reactions that produce small molecules needed for growth, or successive steps needed to break down a small molecule used for nutrients or energy. Molecules of mRNA that code for multiple proteins are known as polycistronic mRNA because the term “cistron” was once widely used to refer to a protein-coding sequence in a gene.
3.4.2 Primary transcripts in eukaryotes undergo several types of chemical modification.
In eukaryotes, the nuclear envelope is a barrier between the processes of transcription and translation. Transcription takes place in the nucleus, and translation in the cytoplasm. The separation allows for a complex chemical modification of the primary transcript, known as RNA processing, which converts the primary transcript into the finished mRNA, which can then be translated by the ribosome.
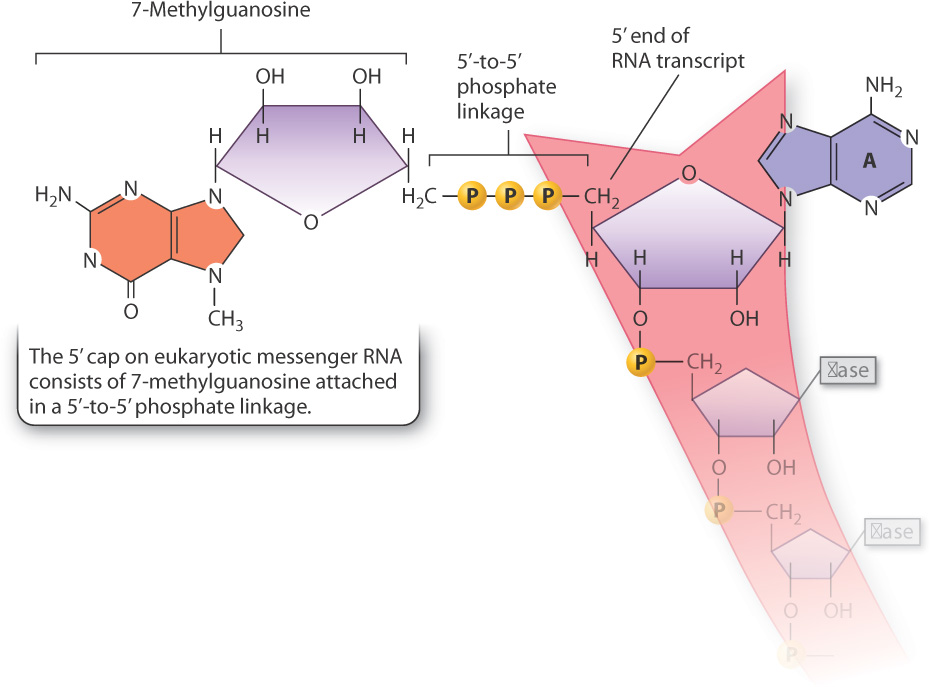
RNA processing consists of three principal types of chemical modification, illustrated in Fig. 3.22. First, the 5′ end of the primary transcript is modified by the addition of a special nucleotide attached in an unusual linkage. This addition is called the 5′ cap, and it consists of a modified nucleotide called 7-methylguanosine. An enzyme attaches the cap to the 5′ end of the primary transcript essentially backwards: in a normal linkage between two nucleotides, the phosphodiester bridge forms between the 5′ carbon of one and the 3′-OH group of the next, but here the cap is linked to the RNA transcript by a triphosphate bridge between the 5′ carbons of both ribose sugars (Fig. 3.23). The 5′ cap is essential for translation because in eukaryotes the ribosome recognizes an mRNA by its 5′ cap. Without the cap, the ribosome would not attach the mRNA and translation would not occur.
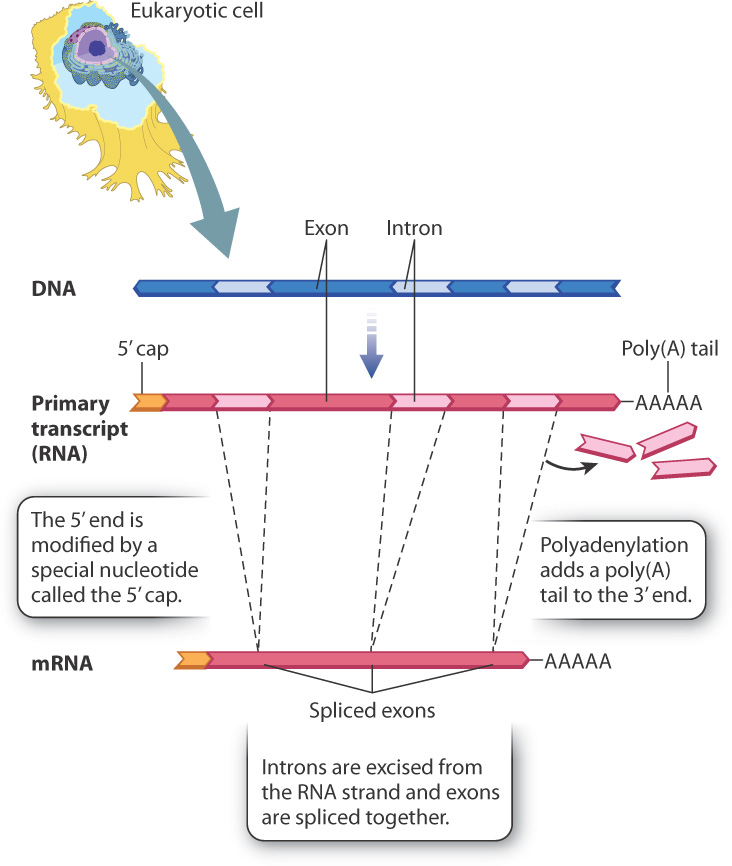
The second major modification of eukaryotic primary transcripts is polyadenylation, the addition of a string of about 250 consecutive A-bearing ribonucleotides to the 3′ end, forming a poly(A) tail (see Fig. 3.22). Polyadenylation plays an important role in transcription termination as well as in the export of the mRNA to the cytoplasm of the cell. In addition, both the 5′ cap and poly(A) tail help to stabilize the RNA transcript. Single-stranded nucleic acids can be unstable and are even susceptible to enzymes that break them down. In eukaryotes, the 5′ cap and poly(A) tail protect the two ends of the transcript and increase the stability of the RNA transcript until it is translated in the cytoplasm.
3-17
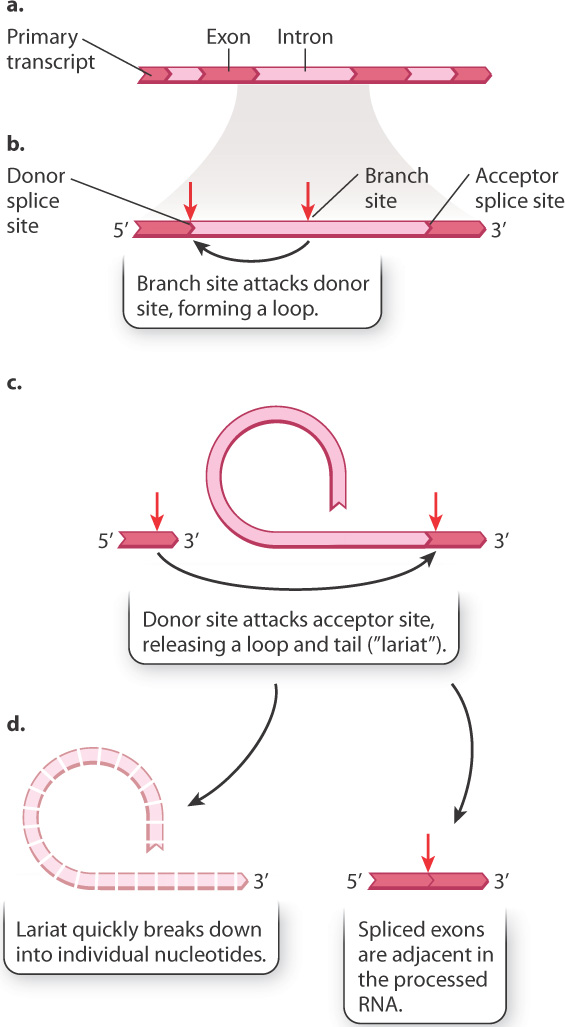
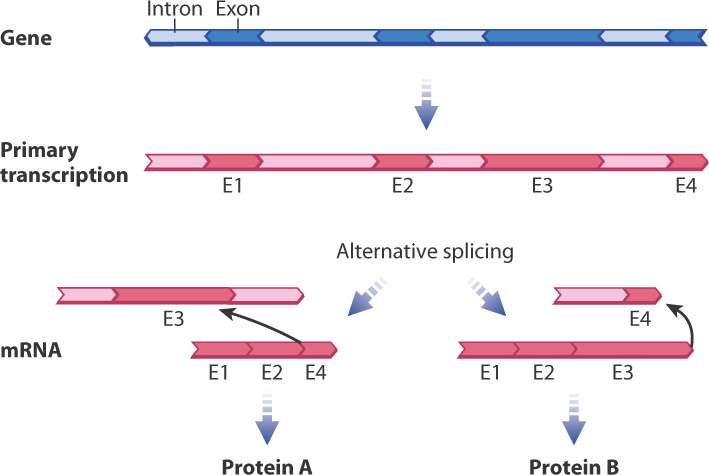
A third modification of the primary transcript is the excision of certain sequences, which are known as introns, from the transcript (see Fig. 3.22). What’s left intact are sequences known as exons. The process of intron removal is known as RNA splicing, which is catalyzed by a complex of RNA and protein known as the spliceosome. The mechanism of splicing is outlined in Fig. 3.24. In the first step, specific sequences near the ends of the intron (Fig. 3.24a) undergo base pairing with RNA molecules in the spliceosome and are brought into close proximity (Fig. 3.24b). The spliceosome enables a reaction that cuts one end of the intron and connects it to a nucleotide near the other, forming a loop and tail called a lariat (Fig. 3.24c). In the next step, the exon on one end of the intron is brought close to the exon at the other end (Fig. 3.24c). These exons are joined and the introns are released (Fig. 3.24d). The lariat making up the intron is quickly broken down into its constituent nucleotides.
About 90% of all human genes contain at least one intron. Although most genes contain 6 to 9 introns, the largest number is 147, found in a muscle gene. Most introns are just a few thousand nucleotides in length, but about 10% are longer than 10,000 nucleotides. The presence of multiple introns in most genes allows for a process known as alternative splicing, in which primary transcripts from the same gene can be spliced in different ways to yield different mRNAs and therefore different protein products (Fig. 3.25). More than 80% of human genes are alternatively spliced. In most cases, the alternatively spliced forms differ in whether a particular exon is or is not removed from the primary transcript along with its flanking introns.
Question Quick Check 4
tPQG28BEUNh6CEH9F4M+IuMMQLam9VByX2PLtdgbrG8QajOxY8cmiKIEqbFKyBYE3vbfBt/2Hz+WZ1Kx2fu9eslvHy5nj3kB3tzkJz7SqN+J9dPJyE9tbpHT9R4eTFwI79WT37vaAWW9REvvfgwyeSLcmQcLgwP5e0tW7Tai/gMJYH81z6t7DpIDR8UPQh/IT815oHeqm3bOeIYfmAfv9mlveYA8eTb65Mfu2IMjte+aNfMpziQ++BGGdbm7ipLjFKnszcZGVOk4MGqPWXHUwKkSPqA1JTLUpDsj5xh+oufjXV6aVTpYlw3knWjcfJMdp8DCDrqUN5+7hTpE6f8NSWuu+CZtn4YGdtLrJL1zNuMV4QW4nkjcMxCykTkjLlz1WbvTDbkD4cjK82Vu1tCxHTp+SfFSpGWGBrsn4vcWe4EMDYQonESY5NFLnadgxFTFGHLaVMnig+JVZQnKKMZEC3w3cSLScwqa1vQjXvyspKpN7saU3CahUfIlnv9WflIXRXMnPAmcv/oth8JPQ7+Ky2eTyzmSPKofDU8TPQkR1xFOol/EDCLzWqRJ8hw+Rp4bnTXU6Q==3-18
3.4.3 Some RNA transcripts are processed differently from protein-coding transcripts and have functions of their own.
Not all primary transcripts are processed into mRNA because not all genes code for proteins. Some RNA transcripts have functions of their own, and many of these transcripts are produced by RNA polymerases other than Pol II. These primary transcripts undergo different types of RNA processing, and their processed forms include such important noncoding RNA types as:
- Ribosomal RNA (rRNA), found in all ribosomes that aid in translation.
- Transfer RNA (tRNA) that carries individual amino acids for use in translation.
- Small nuclear RNA (snRNA), found in eukaryotes and involved in splicing, polyadenylation, and other processes in the nucleus.
- Small, regulatory RNA molecules that can inhibit translation (microRNA or miRNA) or cause destruction of an RNA transcript (small interfering RNA or siRNA).
By far, the most abundant transcripts in mammalian cells are those for ribosomal RNA and transfer RNA. In a typical mammalian cell, about 80% of all of the RNA consists of ribosomal RNA, and another approximately 10% consists of transfer RNA. Why are these types of RNA so abundant? The answer is that they are needed in large amounts to synthesize the proteins encoded in the messenger RNA. The roles of mRNA, tRNA, and rRNA in protein synthesis are discussed in the next chapter.