19.2 MESSENGER RNA TO PHENOTYPE IN EUKARYOTES
In eukaryotes, a processed mRNA must exit the nucleus before the translation step of gene expression can occur. The mRNA migrates to the cytoplasm through one of a few thousand nuclear pores, large protein complexes that traverse the nuclear envelope and regulate the inflow and outflow of macromolecules. Once the mRNA is in the cytoplasm, there are multiple opportunities for gene regulation at the levels of mRNA stability, translation, and protein activity (see Figs. 19.1d–19.1f).
19.2.1 Small regulatory RNAs inhibit translation or promote RNA degradation.
Regulatory RNA molecules known as small regulatory RNAs are among the most exciting recent discoveries in gene regulation. They often work by binding to transcripts and blocking translation. Small regulatory RNAs are of exceptional interest to biologists and drug researchers because their small size allows easy synthesis in the laboratory, and researchers can design their sequences to target transcripts of interest.
One type of small regulatory RNA, microRNA (miRNA), starts out just like the RNA transcribed from protein-coding genes, using the same RNA polymerase for transcription and going through the same processes of capping, splicing, and polyadenylation. But there the similarity ends. In miRNA, the processed RNA folds back upon itself to form one or more hairpin structures, or stems-and-loops, stabilized by base pairing in the stem (Fig. 19.10a). Enzymes specifically recognize processed and folded miRNAs and cleave the stems from the hairpin, then further cleave the stem into small double-stranded fragments typically about 20–25 nucleotide pairs long.
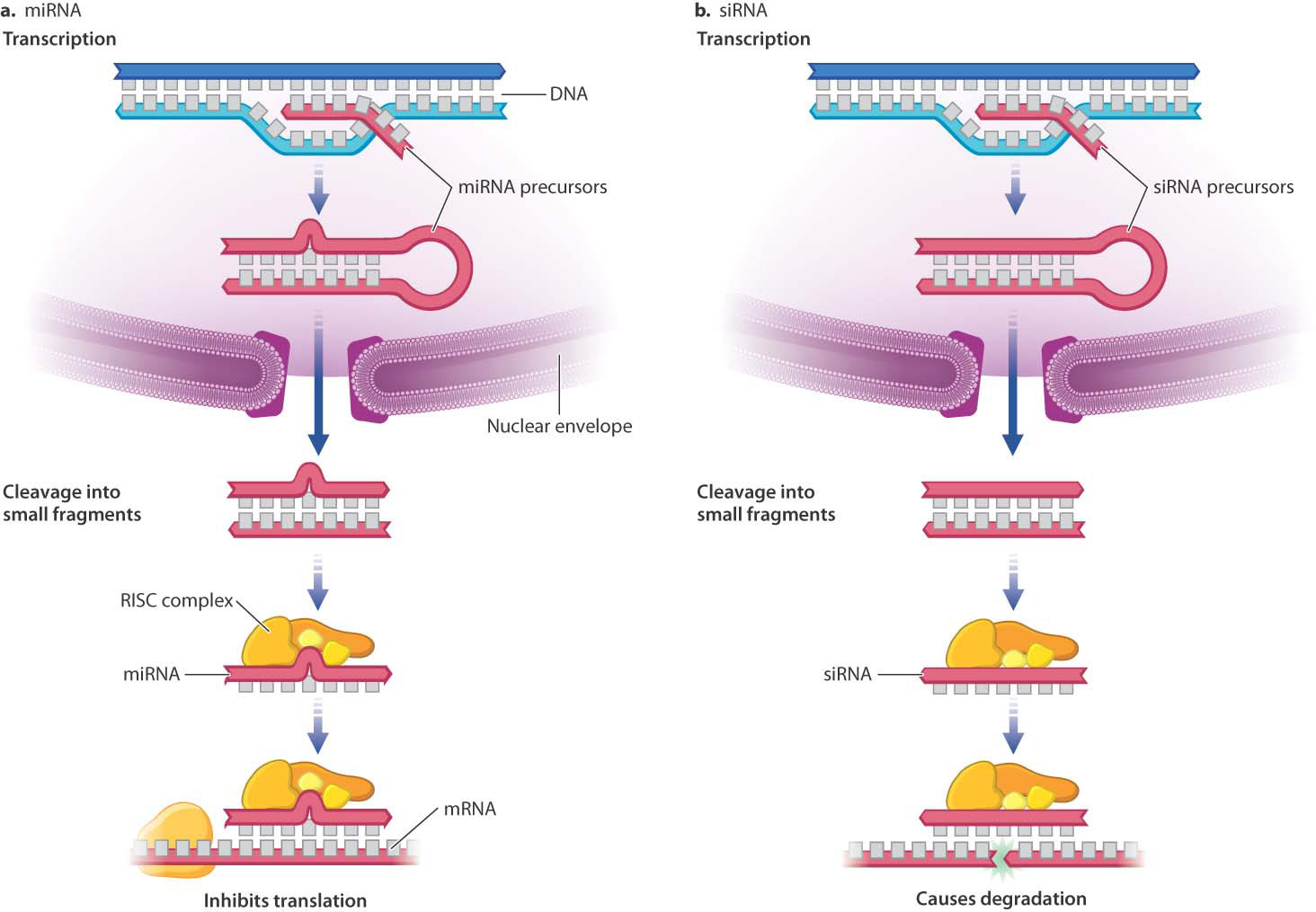
One strand from each RNA fragment is incorporated into a protein complex known as RISC (RNA-induced silencing complex). The small, single-stranded RNA (the miRNA) targets the RISC complex to specific mRNA molecules by base pairing with short regions on the target mRNA (Fig. 19.10a). The miRNA in the RISC complex does not base pair exactly with the target mRNA, but has enough sequence in common that the two strands anneal. Once bound to the target mRNA, the RISC complex inhibits translation. Human chromosomes are thought to encode about 1000 miRNAs, each of which can inhibit translation of tens or hundreds of mRNA molecules. Half or more of human proteins may have their synthesis regulated in part by miRNA.
19-9
A second type of small regulatory RNA is known as small interfering RNA (siRNA). Transcription and processing of siRNA and miRNA are virtually identical, including incorporation into a RISC complex. But unlike miRNAs, which have a few mismatches in their double-stranded region, siRNA molecules have strands that match exactly. RISC complexes containing siRNA cleave the targeted RNA, instead of just binding to the target mRNA (Fig. 19.10b). The cleavage exposes unprotected 5′ and 3′ ends of the single-stranded RNA, which are vulnerable to attack by other nucleases. RISC complexes containing siRNA can lead to degradation of RNA transcripts in the nucleus at any stage during transcription, RNA processing, or export from the nucleus. Regulation by siRNA is widespread in eukaryotes, and is thought to have evolved originally as a defense against viruses and transposable elements.
Question Quick Check 2
B1E5KZ7kURrgNlBfPkxGLIKkEfa2gEx5P5EorHDd4QNOFnuDw1qPGXoCRAFL8/mgCImYx5jq6vIRTtSe19.2.2 Translational regulation controls the rate, timing, and location of protein synthesis.
Translation of mRNA into protein provides another level of control of gene expression. Fig. 19.11 shows the structure of a hypothetical mRNA molecule in a eukaryotic cell and highlights some of the features that help regulate its translation (Chapter 4). Not all mRNA molecules have all the features shown, but all mRNA molecules have a 5′ cap, a 5′ untranslated region (5′ UTR), an open reading frame (ORF) containing the codons that determine the amino acid sequence of the protein, a 3′ untranslated region (3′ UTR), and a poly(A) tail. The 5′ UTR and the 3′ UTR may contain regions that bind with proteins. In some cases, these proteins interact with molecular motors that transport the mRNA to particular regions of the cell. In other cases, the proteins are localized in the cell and repress translation of the mRNAs to which they are bound. By either transport or repression, these proteins cause the mRNA to be translated only in certain places in the cell.
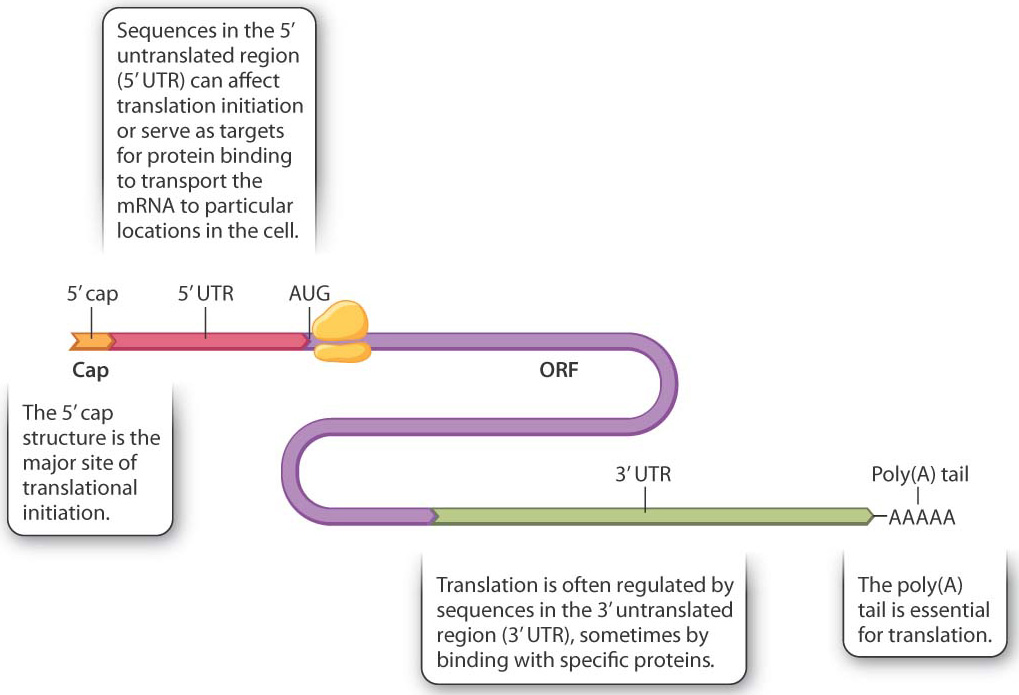
The cap structure is one of the main recognition signals for translation initiation, which requires the coordinated action of about 25 proteins. These are present in most cells in limiting amounts, and so at any one time while some mRNAs from a gene are being translated, other mRNAs transcribed from the same gene may not have a translation initiation complex assembled. Upon formation, the initiation complex moves along the 5′ UTR, scanning for an AUG codon (the initiation codon) to allow the complete ribosome to assemble and begin translation. Although translation initiation is the principal mode of translational regulation, not all mRNA molecules are equally accessible to translation. Among the key variables are the secondary (folded) structure of the 5′ UTR, the distance from the 5′ cap to the AUG initiation codon, and the sequences flanking the AUG initiation codon.
The 3′ UTR and the poly(A) tail are also important in translation initiation. The mRNA forms a loop during initiation and the 3′ end is brought into proximity with the 5′ end. Because of their proximity to the start site for translation, sequences at the 3′ end help regulate translation. In fact, most mRNA sequences that regulate translation are present in the 3′ UTR.
19-10
19.2.3 Protein structure and chemical modification modulate protein effects on phenotype.
Once translation is completed, the resulting protein can alter the phenotype of the cell or organism by affecting metabolism, signaling, or cell structure. After translation, proteins are modified in multiple ways that regulate their structure and function. Collectively, these processes are called posttranslational modification (see Fig. 19.1e). Regulation at this level is essential because some proteins are downright dangerous. For example, proteases such as the digestive enzyme trypsin must be kept inactive until secreted out of the cell. If they were not, their activity would kill the cell itself. These types of protein are often controlled by being translated in inactive forms that are made active by modification after secretion.
Folding and acquiring stability are key control points for some proteins (Chapter 4). While many proteins fold properly as they come off the ribosome, others require help from other proteins, called chaperones, which act as folding facilitators. Correct folding is important because improperly folded proteins may form aggregates that are destructive to cell function. Many diseases are associated with protein aggregates, including Alzheimer’s disease, Huntington’s disease, and mad cow disease.
Posttranslational modification also helps regulate protein activity. Many proteins are modified by the addition of one or more sugar molecules to the side chains of some amino acids. This modification can alter the protein’s folding and stability, or target the molecule to particular cellular compartments. Reversible addition of a phosphate group to the side groups of amino acids such as serine, threonine, or tyrosine is a key regulator of protein activity (Chapter 9). Introduction of the negatively charged phosphate group alters the conformation of the protein, in some cases switching it from an inactive state to an active state and in other cases the reverse. Because the function of a protein molecule results from its shape and charge (Chapter 4), a change in protein conformation affects protein function.
Marking proteins for enzymatic destruction by the addition of chemical groups after translation is also important in controlling their activity. We have already seen how the destruction of successive waves of cyclin proteins helps move the cell through its division cycle (Chapter 11).
19.2.4 How do lifestyle choices affect expression of your personal genome?
Case 3 You, From A to T: Your Personal Genome
What does gene regulation have to do with you? If you examine Fig. 19.1 as a whole and consider the DNA sequence shown as your personal genome, the situation looks pretty grim. You might be led to believe that genes dictate everything, and that biology is destiny. But if you focus on the lower levels of regulation in Fig. 19.1, a different picture emerges. The picture is different because much of the regulation that occurs after transcription (posttranslational modifications, regulation of translation, regulation of RNA stability) is determined by the physiological state of your cells, which in turn is strongly influenced by your lifestyle choices. For example, your cells can synthesize 12 of the amino acids, but if any of these is present in sufficient amounts in your diet, it is absorbed during digestion and not synthesized. The essential amino acid you ingest blocks the synthetic pathway through feedback effects.
The effect of an intervention—genetic or environmental—at any given level can affect regulatory processes at both higher and lower levels. This cascade of regulatory effects in both directions can occur because the expression of any gene is regulated at multiple levels, and because there is much feedback and signaling back and forth between nucleus and cytoplasm. It is because of these feedback and signaling mechanisms that the effects of lifestyle choices can be propagated up the regulatory hierarchy. For example, it has been shown that dietary intake of fats and cholesterol affects not only the activity of enzymes directly involved in the metabolism of fats and cholesterol, but also the levels of transcription of the genes encoding these enzymes by affecting the activity of their regulatory transcription factors. Similarly, lifestyles that combine balanced diets with exercise and stress relief have been shown to increase transcription of genes whose products prevent cellular dysfunction and to decrease transcription of genes whose products promote disease, including cancer.
So far, we have been talking primarily about complex traits of the type discussed in Chapter 18, which are affected by multiple genes and by multiple environmental factors as well as by genotype-by-environment interaction. For example, there are both genetic and environmental risk factors for breast and ovarian cancers, as we have seen. Simple Mendelian traits caused by mutations in single genes, such as cystic fibrosis and alpha-1 antitrypsin (α1AT) deficiency (Chapter 17), are less responsive to lifestyle choices. But even in these cases, lifestyle does matter—for example, people with α1AT deficiency should not smoke tobacco and should avoid environments with low air quality.