20.2 HIERARCHICAL CONTROL OF DEVELOPMENT
During development of a complex multicellular organism, many genes are activated and repressed at different times, thus restricting cell fates. One of the key principles of development is that genes expressed early in an organism’s development control the activation of other groups of genes that act later in development. Gene regulation during development is therefore hierarchical in the sense that genes expressed at each stage in the process control the expression of genes that act later.
20.2.1 Drosophila development proceeds through egg, larval, and adult stages.
The fruit fly Drosophila melanogaster has played a prominent role in our understanding of the genetic control of early development, and in particular the hierarchical control of development. Researchers have isolated and analyzed a large number of mutant genes that lead to a variety of defects at different stages in development. These studies have revealed many of the key genes and processes in development, which are the focus of the following sections.
The major events in Drosophila development are illustrated in Fig. 20.5. DNA replication and nuclear division begin soon after the egg and sperm nuclei fuse (Fig. 20.5a). Unlike in mammalian development, the early nuclear divisions in the Drosophila embryo occur without cell division, and therefore the embryo consists of a single cell with many nuclei in the center (Fig. 20.5b). When there are roughly 5000 nuclei, they migrate to the periphery (Fig. 20.5c), where each nucleus becomes enclosed in its own cell membrane, and together they form the cellular blastoderm (Fig. 20.5d).
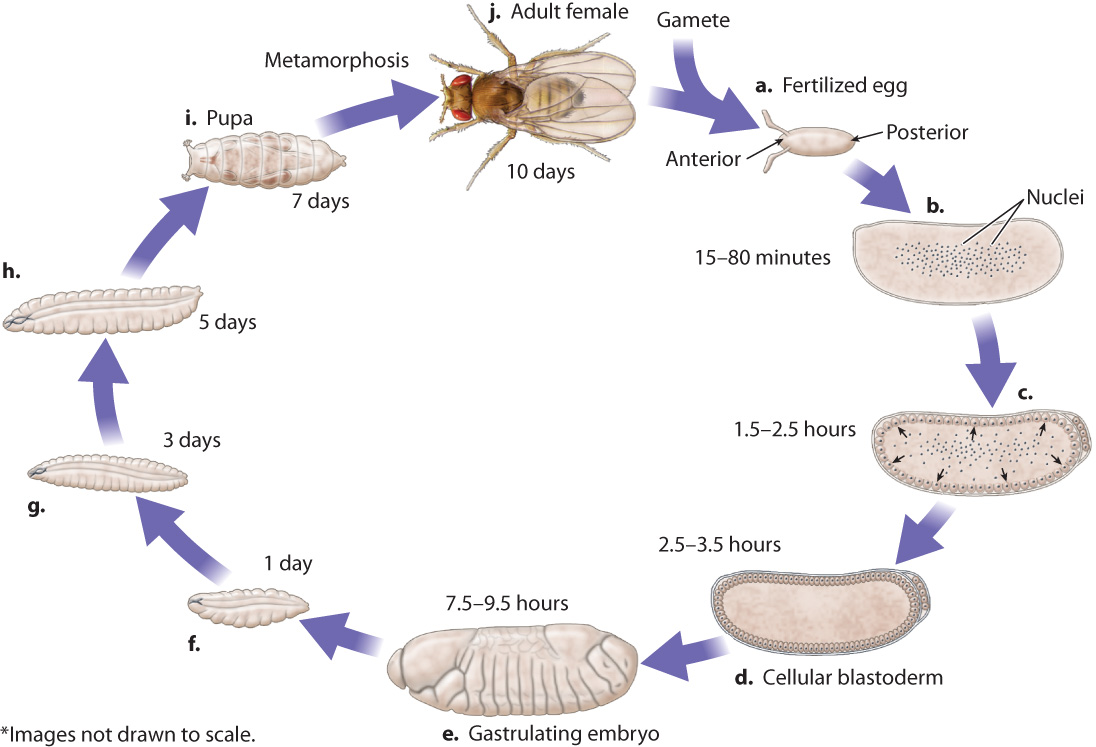
Then begins the process of gastrulation, in which the cells of the blastoderm migrate inward, creating layers of cells within the embryo. As in humans and most other animals (section 20.1 and Chapter 42), gastrulation forms the three germ layers (ectoderm, mesoderm, and endoderm) that differentiate into different types of cell. A Drosophila embryo during gastrulation is shown in Fig. 20.5e. At this stage, the embryo already shows an organization into discrete parts or segments, the formation of which is known as segmentation. There are three cephalic segments, C1–C3 (the term “cephalic” refers to the head); three thoracic segments, T1–T3 (the thorax is the middle region of an insect); and eight abdominal segments (A1–A8). Each of these segments has a different fate in development.
20-6
About one day after fertilization, the embryo hatches from the egg as a larva (Fig. 20.5f). Over the next eight days, the larva grows and replaces its rigid outer shell, or cuticle, twice (Fig. 20.5g and h). After a week of further growth, the cuticle forms a casing—called the pupa—in which the larva is immobilized (Fig. 20.5i), and where it undergoes dramatic developmental changes known as metamorphosis that give rise to the adult fruit fly.
20.2.2 The egg is a highly polarized cell.
How genes control development in Drosophila was inferred from systematic studies of mutants by Christiane Nusslein-Volhard and Eric F. Wieschaus, work for which they were awarded the 1995 Nobel Prize in Physiology or Medicine. One of their findings was that development starts even before a zygote is formed, in the maturation of the oocyte, the unfertilized egg cell produced by the mother. The oocyte, which matures under control of the mother’s genes, is nonetheless important for normal embryonic development. This finding applies not only to insects like Drosophila, but also to many multicellular animals.
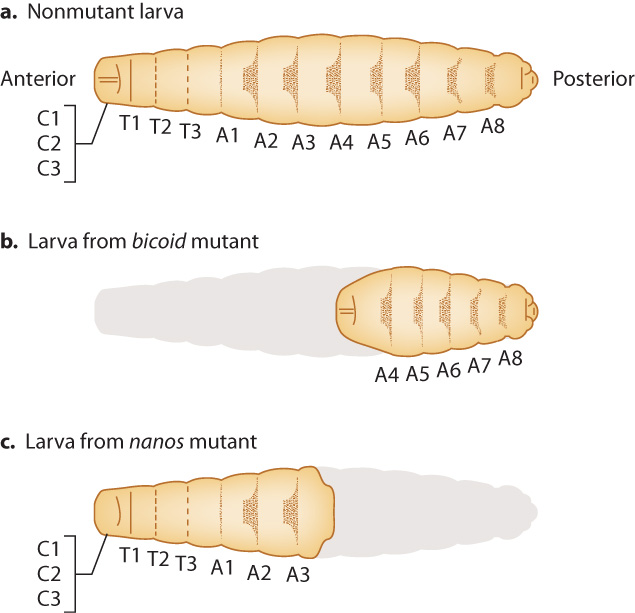
Among the striking mutants Nusslein-Volhard and Wieschaus generated and investigated were ones that significantly affected early development (Fig. 20.6). These defects in very early development can be easily seen by the time the mutants reach the larval stage. In one class of mutants, called bicoid, larvae are missing segments at the anterior end. In another class of mutants, called nanos, larvae are missing segments at the posterior end. The mutant larvae are grossly abnormal and do not survive. Nusslein-Volhard and Wieschaus were able to identify each segment that was missing based on each segment’s distinctive pattern of hairlike projections. In Fig. 20.6, the patterns are shown as dark shapes on each segment.
A distinguishing feature of bicoid and nanos mutants is that the abnormalities in the embryo depend on the genotype of the mother, not the genotype of the embryo. The reason the genotype of the mother can affect the phenotype of the developing embryo is that successful development requires a functioning oocyte. In Drosophila and many other organisms, the composition of the egg includes macromolecules (such as RNA and protein) synthesized by cells in the mother and transported into the egg. If the mother carries mutations in genes involved in the development of the oocyte, the offspring can be abnormal. Genes such as bicoid and nanos that are expressed by the mother but affect the phenotype of the offspring (in this case the developing embryo and larvae) are called maternal-effect genes.
20-7
A normal Drosophila oocyte is highly polarized, meaning that one end is distinctly different from the other. For example, there are gradients of macromolecules that define the anterior–posterior axis of the embryo as well as the dorsal–ventral axis. The best known of these gradients are those of messenger RNAs that correspond to the maternal-effect genes bicoid and nanos. Fig. 20.7a shows the gradient of bicoid mRNA across the oocyte. The bulk of bicoid mRNA comes from the mother, not from the oocyte, and is localized in the anterior of the egg by proteins that attach them to the cytoskeleton.
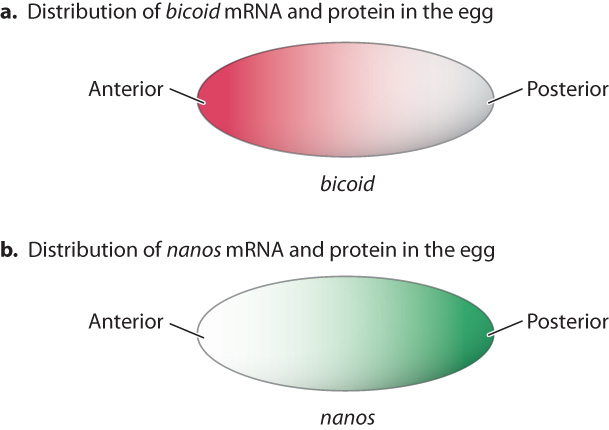
The mRNA corresponding to the maternal-effect gene nanos is also present in a gradient, but its maximum is at the posterior end (Fig. 20.7b). Like bicoid, mRNA for nanos is synthesized by the mother’s cells and then imported into the oocyte. After fertilization, the zygote produces Bicoid and Nanos proteins, and they have concentration gradients resembling those of the mRNAs in the oocyte (Fig. 20.7).
The anterior–posterior axis set up by the gradients of Bicoid and Nanos proteins is reinforced by gradients of two transcription factors called Caudal and Hunchback (Fig. 20.8). Like the mRNAs for Bicoid and Nanos, the mRNAs for Caudal and Hunchback are transcribed from the mother’s genome and transported into the egg. As shown in Fig. 20.8a, the mRNAs for caudal and hunchback are spread uniformly in the cytoplasm of the fertilized egg. However, the mRNAs are not translated uniformly in the egg. Bicoid protein represses translation of caudal, and Nanos protein represses translation of hunchback (Fig. 20.8b). Caudal protein is therefore concentrated at the posterior end and Hunchback protein is concentrated at the anterior end. The expression of Caudal and Hunchback illustrates gene regulation at the level of translation, discussed in Chapter 19. Bicoid protein is also a transcription factor that promotes transcription of the hunchback gene from zygotic nuclei, which reinforces the localization of Hunchback protein at the anterior end.
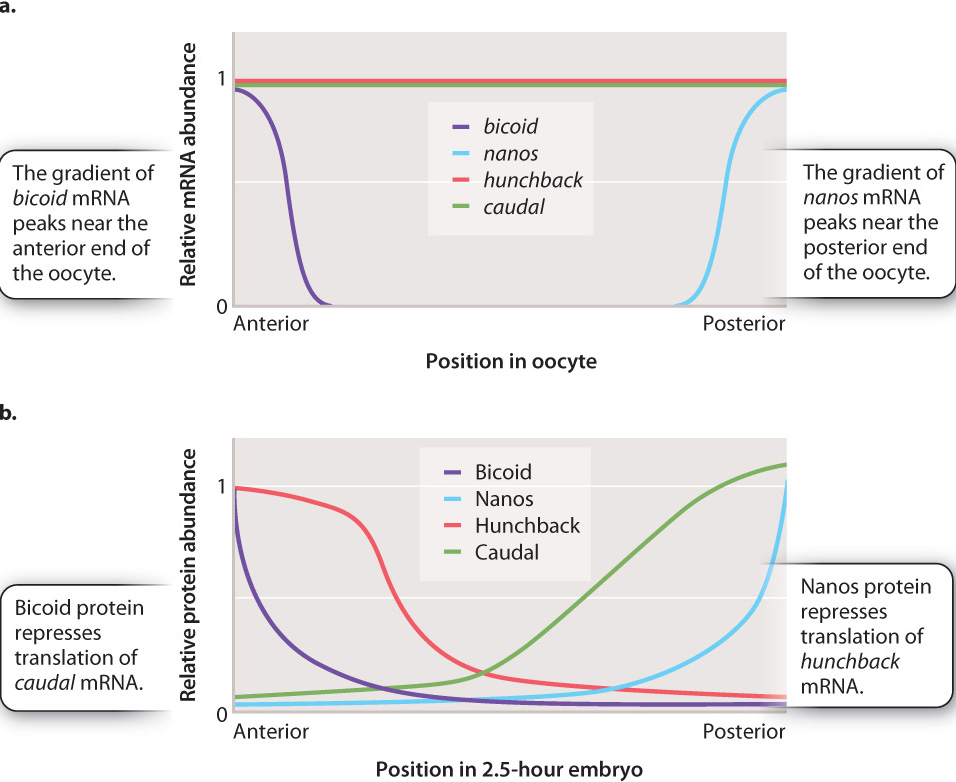
The Hunchback and Caudal gradients set the stage for the subsequent steps in development. The Hunchback transcription factor targets genes needed for the development of anterior structures like eyes and antennae, and Caudal targets genes needed for the development of posterior structures like legs. In this way, genes expressed early in development influence the expression of genes important in later development. Because the products of the bicoid and nanos mRNA are the ones initially responsible for organizing the anterior and posterior ends of the embryo, respectively, mothers that are mutant for bicoid have larvae that lack anterior structures, and mothers that are mutant for nanos have larvae that lack posterior structures.
Question Quick Check 3
fOf6hE1HxERg9WgN08KfSay4Kgi+F60Hz8wlGFiwCMxzxtyWG5v+m+UhBIMn0doJABYTiRVBFirXmThhodjEOu6fKwzg9LdkGyrSR2Im0t8q8q/bO6lKWsgSq6a1oGdQSaD1yYjGJJrcVEK+czITgzY3AlTJWQHyTn4jnhhRUgc=20.2.3 Development proceeds by progressive regionalization and specification.
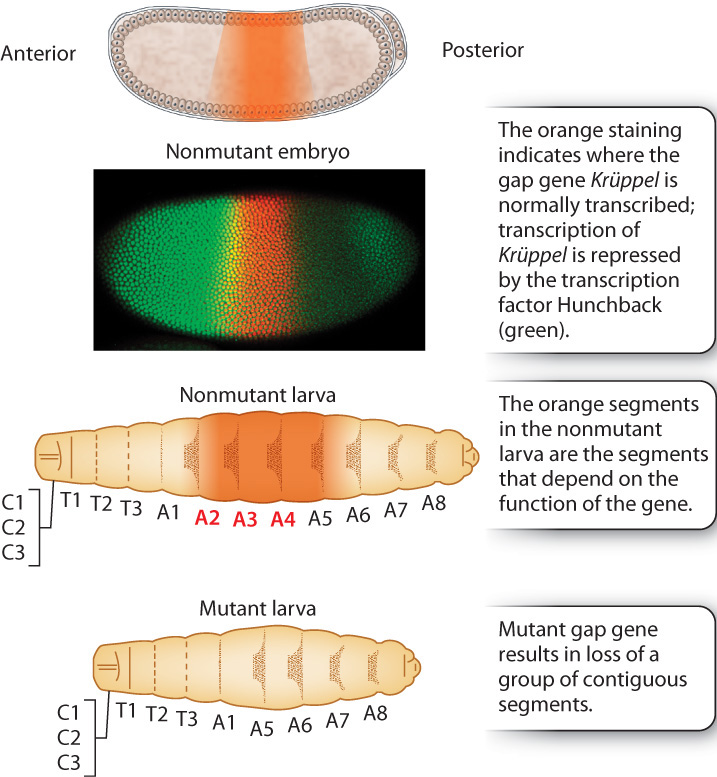
Nusslein-Volhard and Wieschaus also discovered mutants in three other classes of developmental genes. Analysis of these mutants showed that genes controlling development are turned on in groups, and that each successive group acts to refine and narrow the pattern of differentiation generated by previous groups. This, too, is a general principle of development in many multicellular organisms.
The anterior–posterior gradient set up by the maternal-effect genes is first narrowed by genes called gap genes (Fig. 20.9), each of which is expressed in a broad region of the embryo. The name “gap gene” derives from the phenotype of mutant embryos, which are missing groups of adjoining segments, leaving a gap in the pattern of segments. Fig. 20.9 shows the expression pattern of the gap gene Krüppel, which is expressed in the middle region of the embryo. Mutants of Krüppel lack some larval thoracic and abdominal segments. Krüppel is expressed in the pattern shown in Fig. 20.9 because it is under the control of the transcription factor Hunchback, which in this embryo is stained in green. High concentrations of Hunchback repress Krüppel transcription entirely, and low concentrations fail to induce Krüppel transcription. Because Hunchback is present in an anterior–posterior gradient (see Fig. 20.8), the pattern of Hunchback expression means that Krüppel is transcribed only in the middle region of the embryo where Hunchback is present but not too abundant.
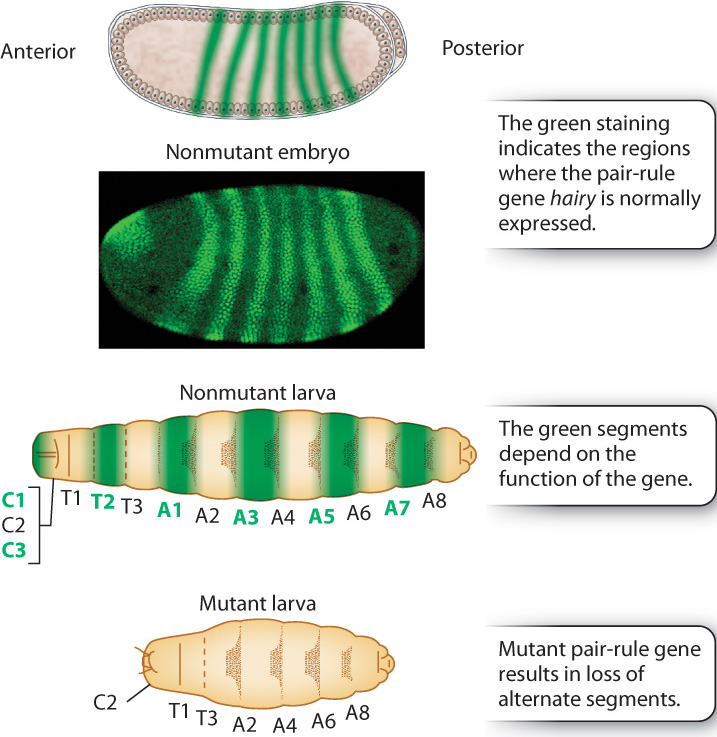
The next level of the regulatory hierarchy is that of pair-rule genes (Fig. 20.10), which receive their name because their mutants lack alternate larval segments. The example in Fig. 20.10 is hairy, whose mutants lack the odd-numbered thoracic segments and the even-numbered abdominal segments. The pair-rule genes help to establish the uniqueness of each of seven broad stripes across the anterior–posterior axis.
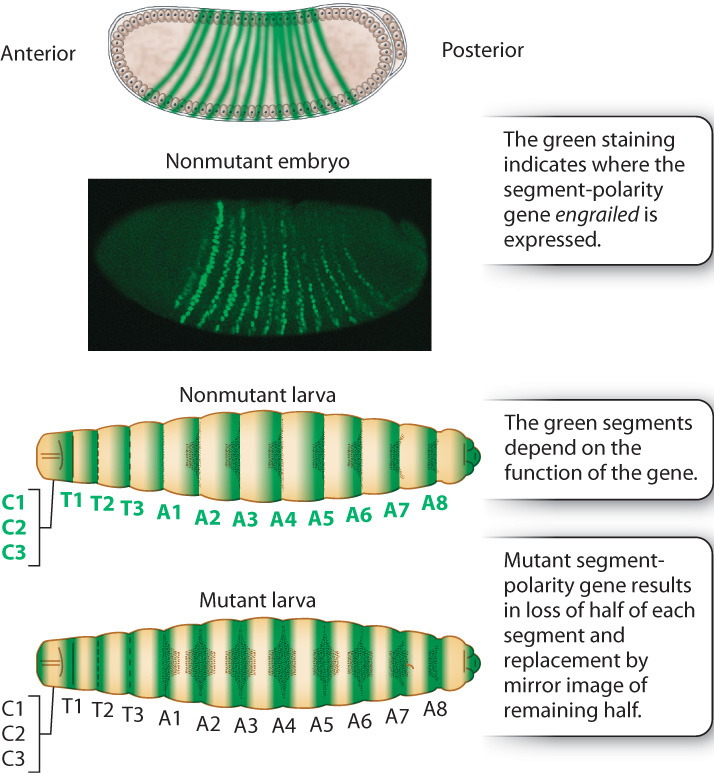
The pair-rule genes in turn help to regulate the next level in the segmentation hierarchy, which consists of segment-polarity genes (Fig. 20.11). The segment-polarity genes refine the 7-striped pattern still further into a 14-striped pattern. Each of the 14 stripes has distinct anterior and posterior ends determined by the segment-polarity genes. Embryos with mutations in segment-polarity genes lose this anterior–posterior differentiation, with the result that the anterior and the posterior halves of each segment are mirror images. The example in Fig. 20.11 is engrailed, which eliminates the posterior pattern element in each stripe and replaces it with a mirror image of the anterior pattern element.
20-9
Question Quick Check 4
an9p8T23wo/JEx06rddVIc6q/HAABs5/lNtvS47SEzh1tx+OL/w+KFYiopkllho0yhHtq5DwTxSIBo+9KHzwWOR6MHg2xCeAcwSb0HhSJa/mxCAjnZeZrzGur8FrlTpS+jll8eVoLD6uOic1Io2EK7ntahRl71k4gGMFygXP/j7uVsx5kBFF0hZy/B4=20.2.4 Homeotic genes determine where different body parts develop in the organism.

Together, the segment-polarity genes and certain genes expressed earlier in the hierarchy control the pattern of expression of another set of genes called homeotic genes. Originally discovered in Drosophila, homeotic genes encode some of the most important transcription factors in animal development. A homeotic gene is a gene that specifies the identity of a body part or segment during embryonic development. For example, homeotic genes instruct the three thoracic segments (T1, T2, and T3) to each develop a set of legs, and the second thoracic segment (T2) to develop wings.
Two classic examples of the consequences of mutations in homeotic genes in Drosophila are shown in Fig. 20.12. Fig. 20.12a shows what happens when the homeotic gene Antennapedia, which specifies the development of the leg, is inappropriately expressed in anterior segments. The mutation causes legs to grow where antennae usually would. Similarly, a mutation in the homeotic gene Bithorax results in the transformation of thoracic segment 3 (T3) into thoracic segment 2 (T2), so that the fruit fly has two T2 segments in a row. As shown in the Fig. 20.12b, the result is a fruit fly with two complete sets of wings.
20-10
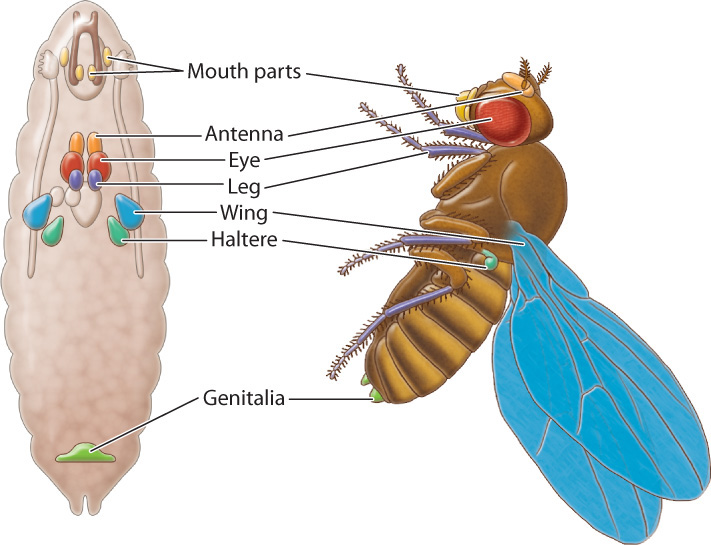
In Drosophila, the adult body parts like legs, antennae, and wings are formed from organized collections of tissue located throughout the larval body (Fig. 20.13). The development of these tissues and their metamorphosis into the adult body parts are regulated by the homeotic genes. First expressed at about the same time as the segment-polarity genes (see Fig. 20.11), the homeotic genes continue to be expressed even after the genes that regulate early development have shut down. Their continuing activity is due to the presence of chromatin remodeling proteins that keep the chromatin physically accessible to the transcription complex (Chapter 19).
Homeotic genes encode transcription factors. The DNA-binding domain in the homeotic proteins is a sequence of 60 amino acids called a homeodomain, whose sequences are very similar from one homeotic protein to the next. The homeodomain is specified by a DNA sequence within the homeotic genes called the homeobox (the name is the origin of the term Hox gene, which is often used as a synonym for “homeotic gene”).
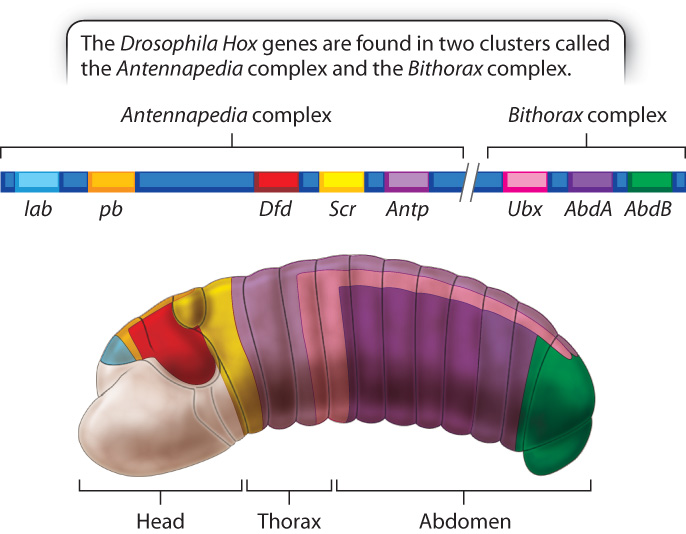
The Drosophila genome contains eight Hox genes comprising two distinct clusters, the Antennapedia complex and the Bithorax complex (Fig. 20.14). The genes are arranged along the chromosome in the same order as their products function in anterior–posterior segments along the embryo. In addition, the timing of their expression corresponds to their order along the chromosome and location of expression, with genes that are expressed closer to the anterior end turned on earlier than genes that are expressed closer to the posterior end. The correlation among linear order along the chromosome, anterior–posterior position in the embryo, and timing of expression is observed in Hox clusters in almost all organisms studied.
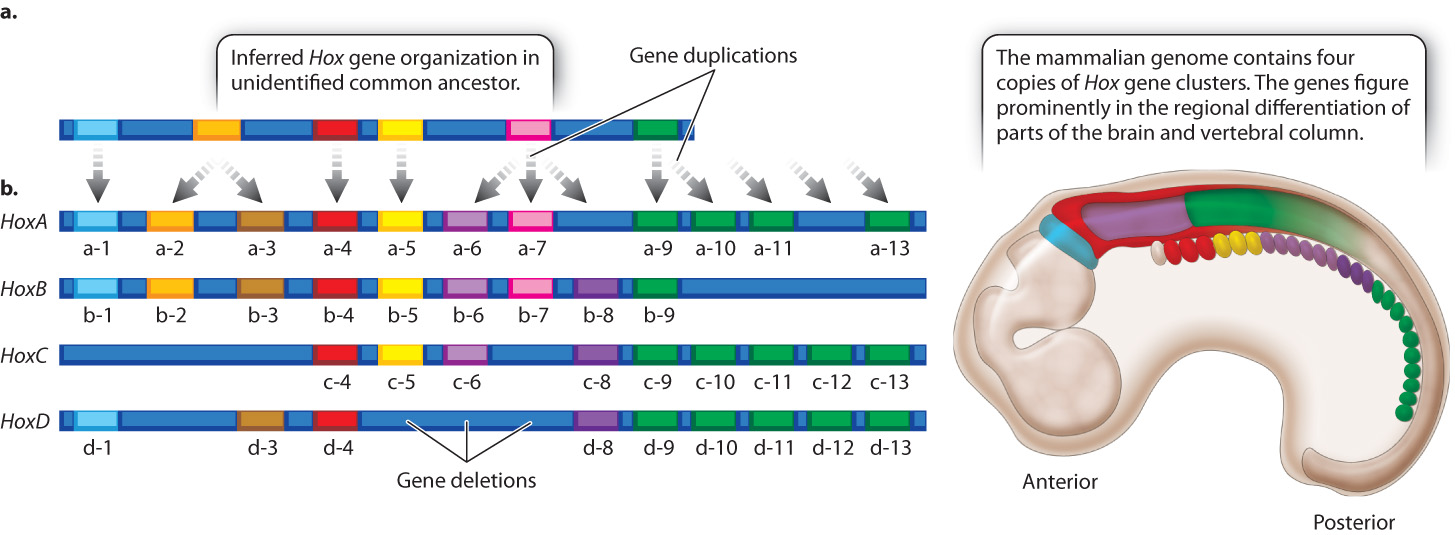
Because the amino acid sequences of the homeodomains of Hox gene products are very similar from one organism to the next, Hox gene clusters have been identified in a wide variety of animals with bilateral symmetry (organisms in which both sides of the midline are mirror images), from insects to mammals. Comparison of the number and types of Hox genes in different species supports the hypothesis that the ancestral Hox gene cluster had an organization very similar to what we now see in most organisms with Hox gene clusters. In its evolutionary history, the vertebrate genome underwent two whole-genome duplications; hence, vertebrates have four copies of the Hox gene cluster (Fig. 20.15).
Unlike the Hox genes in Drosophila, mammalian Hox genes do not specify limbs but are important in the embryonic development of structures that become parts of the hindbrain, spinal cord, and vertebral column (Fig. 20.15). As in Drosophila, the genes in each cluster are expressed according to their linear order along the chromosome, which coincides with the linear order of regions the genes affect in the embryo. Each gene helps to specify the identity of the region in which it is expressed. Many of the genes in the mammalian Hox clusters have redundant or overlapping functions so that learning exactly what each gene does continues to be a research challenge.
20-11