21.6 MOLECULAR EVOLUTION
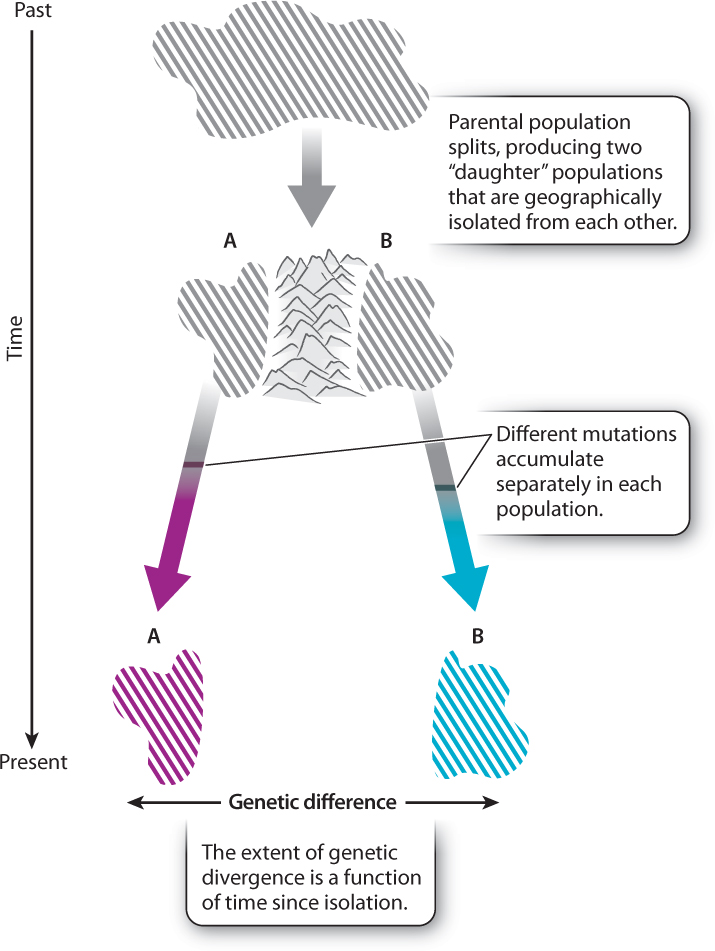
How do DNA sequence differences arise among species? Imagine starting with two pairs of identical twins, one pair male and the other female. Now we place one member of each pair together on either side of a mountain range (Fig. 21.13). Let’s assume the mountain range completely isolates the couples on each side. What, in genetic terms, will happen over time? The original pairs will found populations on each side of the mountain range. The genetic starting point is, in each case, exactly the same, but, as the generations tick by, differences will accumulate between the two populations. Over time, mutations will occur in one population that will not have arisen in the other population, and vice versa. Because mutations occur at a low rate and are scattered randomly throughout the genome, the chance that both populations acquire the same mutations is practically zero.
A mutation in either population ultimately suffers one of three fates: It goes to fixation (either through genetic drift or through positive selection); it is maintained at intermediate frequencies (by balancing selection); or it is eliminated (either through natural selection or genetic drift). Different mutations will be fixed in each population. When we come back thousands of generations later and sequence the DNA of our original identical individuals’ descendants, we will find that many differences have accumulated. The populations have diverged genetically. What we are seeing is evidence of molecular evolution.
Species are the biological equivalents of islands because they, too, are isolated. They are genetically isolated because, by definition, members of one species cannot exchange genetic material with members of another (Chapter 22). The amount of time that two species have been isolated from each other is the time since their most recent common ancestor. Thus, humans and chimpanzees, whose most recent common ancestor lived about 6-7 million years ago, have been isolated from each other for about 6-7 million years. Mutations arose and were fixed in the human lineage over that period; mutations, usually different ones, also arose and were fixed in the chimpanzee lineage over the same period. The result is the genetic difference between humans and chimpanzees. If we sequence a particular gene for both species, we find a number of sequence differences.
21.6.1 The extent of sequence difference between species is a function of the time since the species diverged.
The extent of genetic difference, or genetic divergence, between two species is a function of the time they have been genetically isolated from each other. The longer they have been apart, the greater the opportunity for mutation and fixation to occur in each population. This correlation between the time two species have been evolutionarily separated and the amount of genetic divergence between them is known as the molecular clock.
For a clock to function properly, it not only needs to keep time, but it also needs to be set. We set the clock using dates from the fossil record. For example, in a 1967 study, Vince Sarich and Allan Wilson deduced from fossils that the lineages that gave rise to the Old and New World monkeys separated about 30 million years ago. Finding that the amount of genetic divergence between humans and chimpanzees was about one-fifth of that between Old and New World monkeys, they concluded that humans and chimpanzees had been separated one-fifth as long, or about 6 million years. Although this is the generally accepted number today, Sarich and Wilson’s result was revolutionary at the time, when it was thought that the two species were separated for as long as 25 million years.
21.6.2 The rate of the molecular clock varies.
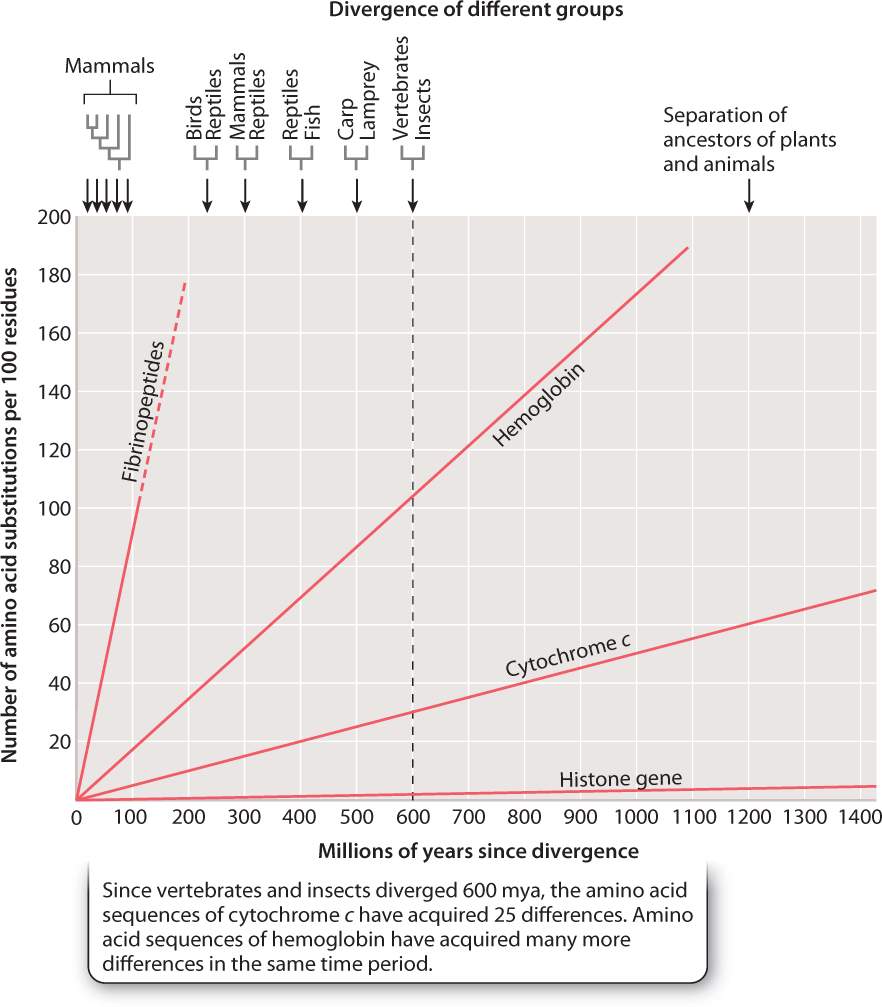
Molecular clocks can be useful for dating evolutionary events like the separation of humans and chimpanzees. However, because the rates of molecular clocks vary from gene to gene, clock data should be interpreted cautiously. These rate differences can be attributed largely to differences in intensity of negative selection (which results in the elimination of harmful mutations) among different genes. The slowest molecular clock on record belongs to the histone genes, which encode the proteins around which DNA is wrapped to form chromatin (Chapter 3). These proteins are exceptionally similar in all organisms; only 2 amino acids (in a chain of about 100) distinguish plant and animal histones. Plants and animals last shared a common ancestor more than 1 billion years ago, which means, because each evolutionary lineage is separate, that there have been at least 2 billion years of evolution since they were in genetic contact. And yet, in all that time, the histones have hardly changed at all. Almost any amino acid change fatally disrupts the histone protein, preventing it from carrying out its proper function. Negative selection has thus been extremely effective in eliminating just about every amino acid-changing histone mutation over 2 billion years of evolution. The histone molecular clock is breathtakingly slow.
21-16
Other proteins are less subject to such rigorous nagative selection. Occasional mutations may therefore become fixed, either through drift (if they are neutral) or selection (if beneficial). The extreme case of a fast molecular clock is that derived from a pseudogene, a gene that is no longer functional. Because all mutations in a pseudogene are by definition neutral—there is no function for a mutation to disrupt, so a mutation is neither deleterious nor beneficial—we expect to see a pseudogene’s molecular clock tick at a very fast rate. In the histone genes, virtually all mutations are selected against, constraining the rate of evolution; in pseudogenes, none are. Fig. 21.14 vividly shows the varying rates of the molecular clock for different genes.
In the next chapter, we will see how genetic divergence between populations can lead to the evolution of new species.