43.2 ADAPTIVE IMMUNITY: B CELLS, ANTIBODIES, AND HUMORAL IMMUNITY
The innate immune system can react to diverse pathogens and has the capacity to distinguish self (host cells) from non-self (foreign cells). The adaptive immune system shares these features, and has two additional features not present in the innate immune system. The first is specificity: The adaptive immune system targets responses to specific pathogens. As a result, it is often more effective at eliminating a pathogen. The second is memory: The adaptive immune system remembers past infections and mounts a stronger response on re-exposure. How can the adaptive immune system respond to a single pathogen among the diversity of pathogens that it meets? How is immunological memory achieved? We explore answers to these questions in the next two sections.
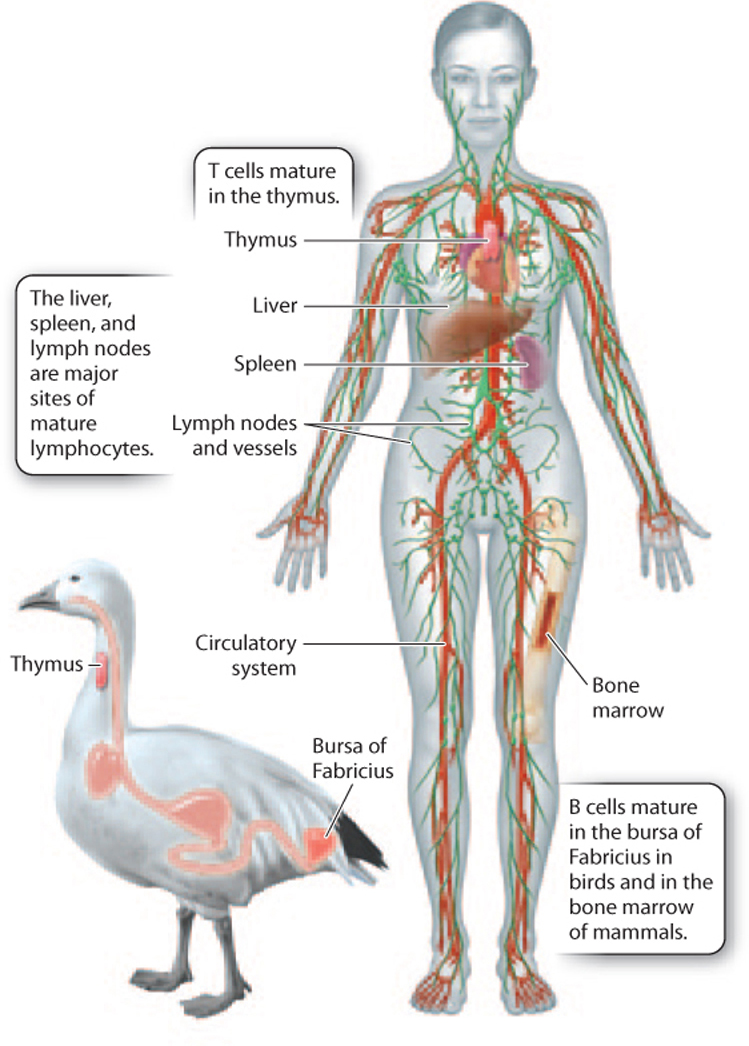
While many cells and tissues play a role in adaptive immunity, two types of cell are particularly important: B lymphocytes, or B cells, and T lymphocytes, or T cells (see Fig. 43.3). B and T cells are named for their site of maturation. In birds, B cells mature in the bursa of Fabricius, an organ of the lymphatic system located off the digestive tract (Fig. 43.8). In mammals, B cells mature in the bone marrow. T cells mature in the thymus, an organ of the immune system located just behind the sternum, or breastbone. B and T cells circulate in blood and lymphatic vessels, and also can be found in the spleen, liver, and lymph nodes (Fig. 43.8). We begin with a discussion of the role of B cells in adaptive immunity.
43.2.1 B cells produce antibodies.
One of the hallmarks of adaptive immunity is the ability to target specific pathogens. But there are a tremendous number of different viruses, bacteria, single-celled eukaryotes, fungi, and worms that can lead to infection. How does the adaptive immune system recognize each one specifically? The specificity of the adaptive immune system is in part the result of antibodies produced by B cells.
An antibody is a large protein that carries sugar molecules attached to some amino acids (hence it is a glycoprotein). An antibody binds to foreign molecules that occur naturally on or in microorganisms and participate in normal cellular functions. Such a molecule is termed an antigen, which is a molecule that binds to and leads to the production of antibodies. (The word “antigen” is a contraction of “antibody” and “generator.”) The great diversity of antigens on different microorganisms is matched by a great diversity of antibodies. It is estimated that humans produce approximately 10 billion different antibodies, each with the ability to recognize one or a few antigens on microorganisms.
Antibodies can be found on the surface of B cells or free in the blood or tissues. Collectively, antibodies make up a significant portion of the proteins present in the blood, typically about 20%. Because antibodies are present in the blood, tissue fluid, and secretions, which used to be called humors, this part of the adaptive immune system is sometimes called humoral immunity.
43-8
Antibodies have a distinctive structure that reflects their specificity for a given antigen. The simplest antibody molecule, shown in Fig. 43.9, is a Y-shaped protein made up of four polypeptide chains—two identical light (L) chains and two identical heavy (H) chains, so called because of their relative molecular weights. Covalent disulfide bonds hold the four chains together.
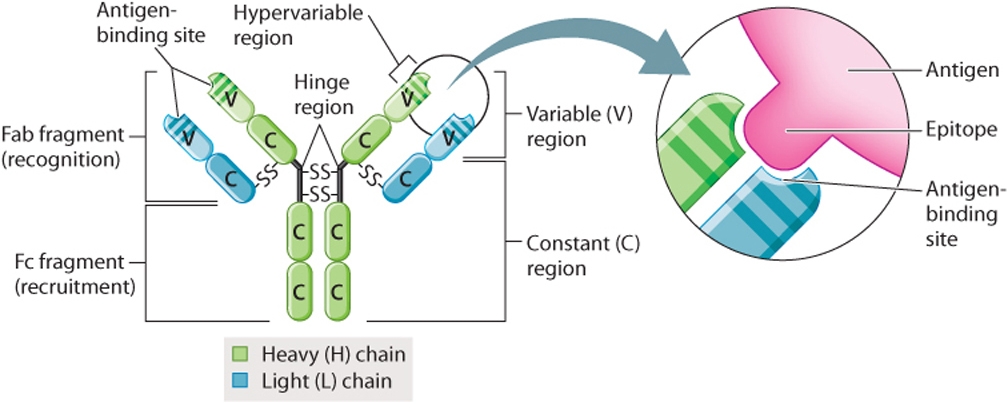
The light and heavy chains are further subdivided into variable (V) and constant (C) regions (Fig. 43.9). Within the variable regions of the L and H chains are regions that are even more variable, which are called hypervariable regions. Together, these hypervariable regions interact in a specific manner with a portion of the antigen called the epitope. Each of the billions of different antibodies has a distinct set of hypervariable regions that recognizes a unique epitope.
Binding of antibodies to antigens is the first step in recognition and removal of microorganisms. Binding alone is sometimes enough to disable a microorganism. In these cases, binding can lead to precipitation of the antibody–antigen complex (agglutination). More often, the microorganism is destroyed by a different component of the immune system, for example, by a phagocyte or the complement system. In this case, the function of an antibody is to recognize the pathogen, then recruit other cells of the immune system or activate the complement system. These latter functions are handled by a different part of the antibody from recognition. This can be demonstrated by treatment with proteolytic enzymes (enzymes that cut proteins at specific locations). For example, one type of enzyme breaks the molecule at the hinge region, producing two Fab fragments and one Fc fragment (Fig. 43.9). (The letters “ab” stand for “antigen-binding”; “c” stands for “crystallizing.”) Each Fab fragment has an antigen-recognition site, and the Fc fragment activates the complement system and binds to cell-surface receptors of other cells of the immune system.
43.2.2 Mammals produce five classes of antibody with different biological functions.
Antibodies are members of a family of proteins with common structural features. As a group, they are called immunoglobulins (Ig). There are five classes of immunogloblulin—IgG, IgM, IgA, IgD, and IgE—each with a different function (Fig. 43.10). These classes are defined by their heavy chains, which differ in the amino acid sequences of their constant regions. There are also two types of light chain. These two types of light chain occur in all five classes of antibody, but any given antibody contains only one type of light chain.
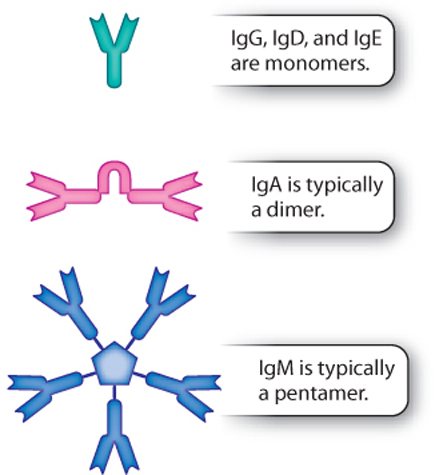
IgG is by far the most abundant of the five antibody classes. It is the Y-shaped antibody depicted in Figs. 43.9 and 43.10. IgG circulates in the blood and is particularly effective against bacteria and viruses. In addition, IgG is the only class of antibody that can cross the placenta because of the presence of receptors on placental cells for the Fc portion of IgG. The ability of IgG to cross the placenta provides protection to the developing fetus.
IgM is a pentamer in mammals and a tetramer in fish. (A pentamer is a molecule made of five monomers; a tetramer is a molecule made of four monomers.) The individual units—the monomers—are linked by a joining chain (Fig. 43.10). IgM also exists as a monomer on the surface of B cells. IgM is particularly important in the early response to infection (discussed below) and is very efficient in activating the complement system and stimulating an immune response.
43-9
IgA is usually a dimer consisting of two antibody molecules linked by a joining chain (Fig. 43.10). It is the major antibody on mucosal surfaces, such as those of the respiratory, gastrointestinal, and genitourinary tracts. It helps to protect mucous membranes from infection. It is also present in secretions, including tears, saliva, and breast milk.
IgD and IgE, like IgG, are monomers. IgD is typically found on the surface of B cells. This immunoglobulin helps initiate inflammation. IgE plays a central role in allergies, asthma, and other immediate hypersensitivity reactions, which are characterized by a heightened or an inappropriate immune response to common antigens. Immediate hypersensitivity reactions result from the binding of the Fc portion of IgE to receptors on the surface of mast cells and basophils. Binding leads to the release of histamine and cytokines, resulting in inflammation that can be life threatening.
The five classes of antibody are also called isotypes. B cells can change the class of antibody they make, a process known as isotype or class switching, discussed below.
43.2.3 Clonal selection is the basis for antibody specificity.
A central question in immunology is how antibody specificity is achieved. There are seemingly limitless types of antigen that an organism might come across. How can B cells generate antibodies that interact only with specific antigens among this tremendous diversity?
Two alternative hypotheses were suggested to explain antibody specificity. In 1900, Paul Ehrlich suggested that there exists a large pool of B cells, each with a different antibody on its surface. The antigen binds to one or a few of these antibodies, stimulating the B cell to divide. In this model, specificity is achieved before any exposure to the antigen. Ehrlich’s model drew on concepts from chemistry such as the binding of an enzyme to a substrate in the manner of a lock and key (Chapter 6). Similarly, the antigen and antibody interact in a specific way.
Later, in the early 1900s, a different hypothesis was suggested. This hypothesis proposed that the antigen instructs the antibody to fold in a particular way so that the two interact in a specific manner. Here, the antigen plays a more active role in the process and antibody specificity is determined only after interaction with the antibody.
Which of these two hypotheses is correct? Does the antigen select a B cell with a preexisting surface antibody with which it interacts specifically, as in the first hypothesis, or does it instruct the B cell to make a specific antibody, as in the second?
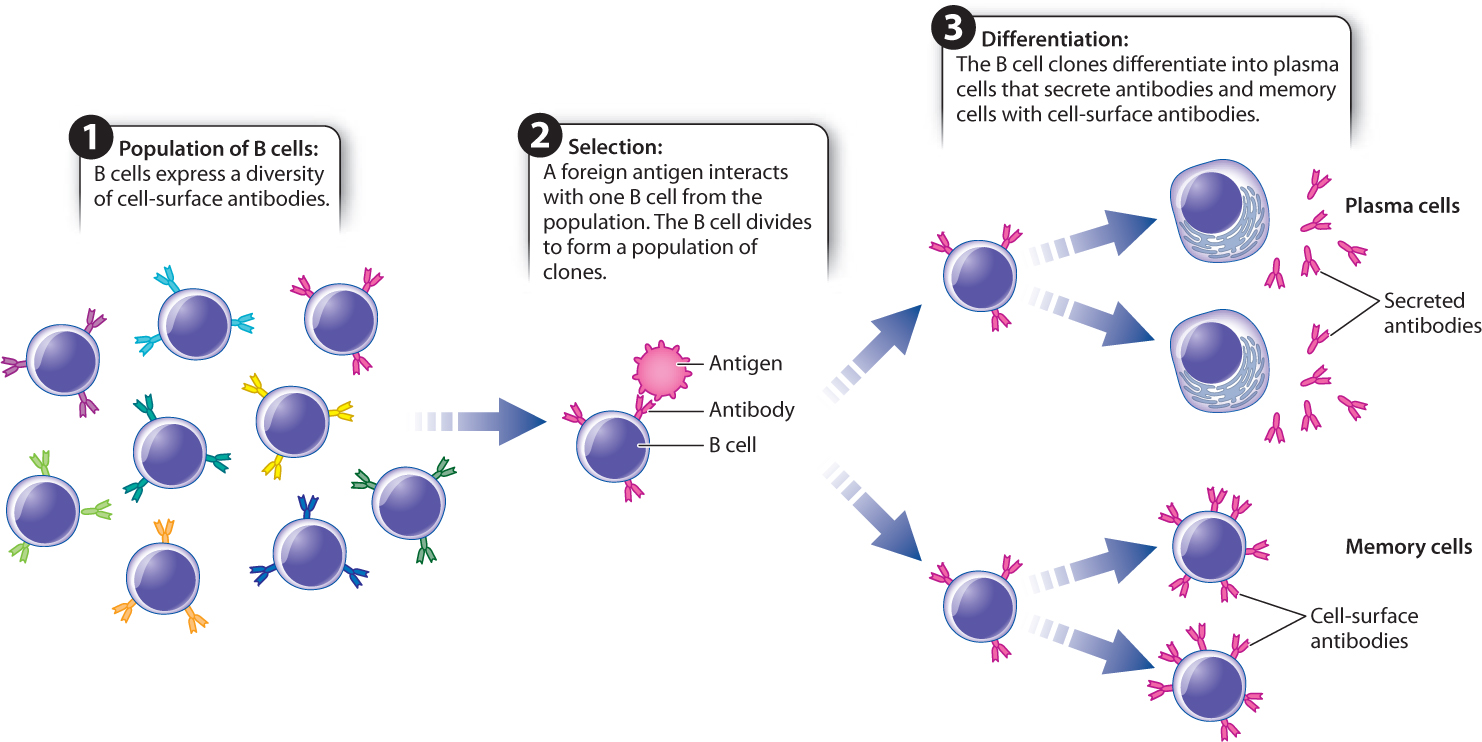
Experimental data on the genetic basis for antibody diversity (described below) provided strong evidence for the first hypothesis. Now called clonal selection, it is one of the central principles of immunology (Fig. 43.11). Every individual has a very large pool of B cells, each with a different antibody on its cell surface. Collectively, these antibodies recognize a great diversity of antigens, but each individual antibody can recognize only one antigen or a few antigens. An antigen interacts with the B cell, or a small set of B cells, that has the best fit between the antigen and the membrane-bound antibody. Binding leads the B cell to divide and differentiate into two types of cell. Plasma, or effector, cells secrete antibodies. Memory cells are long-lived cells that contain membrane-bound antibody having the same specificity as the parent cell. Since the resulting population of B cells arose from one B cell, they are clones of one another. Almost counterintuitively, antibody specificity is achieved before any exposure to the antigen.
43-10
43.2.4 Clonal selection also explains immunological memory.
Clonal selection provides a mechanism for explaining antibody specificity. It also explains the second feature of the adaptive immune system: the ability to remember past infections—that is, to react more vigorously on re-exposure. The first encounter with an antigen leads to a primary response (Fig. 43.12). In this response, there is a short lag before antibody is produced. The lag is the time required for B cells to divide and form plasma cells. The plasma cells secrete antibodies, typically IgM. The level of IgM in the blood increases, peaks, and then declines.
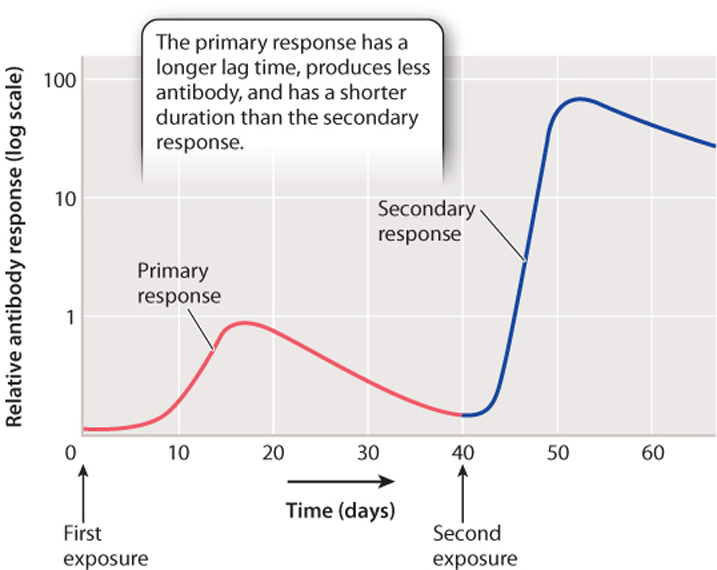
On re-exposure to the same antigen, even months or years after the first exposure, there is a secondary response, which is quicker, stronger, and longer than the primary response (Fig. 43.12). During this response, the antibodies are released by memory B cells. These cells respond more quickly to antigen exposure than naïve B cells (B cells that haven’t been previously exposed to antigen). In addition, the pool of memory B cells is larger than the pool of naïve B cells for a given antigen, explaining why the secondary response produces more antibodies for a longer period of time. IgG is the most abundant antibody in the secondary response.
It is the presence of long-lived memory cells produced following the primary response that provides natural, long-lasting immunity after an infection such as chicken pox. We have learned to take advantage of immunological memory in vaccination. An antigen from a pathogen is deliberately given to a patient in a vaccine to induce a primary response but not the disease, thereby providing future protection from infection.
Until the early 1700s, it was common practice to inoculate people with smallpox in an effort to induce a mild disease and prevent a more severe, even lethal, infection. However, the English scientist Edward Jenner is usually credited with the first demonstration of the protective effects of vaccination. It was well known that milkmaids, exposed to the relatively benign cowpox virus from milking cows, were immune to the related but more deadly smallpox virus. In 1796, Jenner tested the consequences of exposure to cowpox by deliberately inoculating a young boy with cowpox and demonstrating that he became immune to smallpox. In fact, the modern word “vaccine” is derived from the Latin vacca, which means “cow.”
Today, vaccines take many forms: They can be a protein or a part of a protein from the pathogen, a live but weakened form of the pathogen, or a killed pathogen. They are among the most effective public health measures ever developed.
Question Quick Check 2
7kkP2f4/+qz05XOWYkSvzOr0oiw705H/e3ImhYAr0BWEjyGrnYJIzU4C7BhtRzCvRHIIVwvPO9BGxpc05iyY/rVT50FcXeo25JQICyo5J5IOeRS59A/2JD/tpsQhmzaJsJBVNpqYGmNx0+mmtvn5CX/+0YFqlt2cCW+UzXkl51ZW75GS/aWCeC9EtbZ6UL+ZV9JRXE4aEl3UOptiXoVVROGRdSrkdmnCI88SInRMdTKzJk3N4E99DRcZaW/23MI77ZdfZdpDiHFWAEyF9le38+dHCLdwGUayDnW5KbeBnu0SD1eogjvo6pBZiGM=43.2.5 Genomic rearrangement creates antibody diversity.
We pointed out earlier that antibody specificity is determined before exposure to the antigen. In other words, clonal selection requires a mechanism for producing a great diversity of antibodies. How is this diversity achieved? One possibility is that each antibody is encoded by a separate gene. However, this cannot be the case, as the number of antibodies produced by an organism far exceeds the total number of genes in the genome. For example, the human genome consists of about 25,000 protein-coding genes, but humans can produce about 10 billion different antibodies.
In 1965, American biologists William Dreyer and J. Claude Bennett suggested a novel hypothesis to explain antibody diversity. As they put it, this hypothesis was “radically different from anything found in modern molecular genetics.” They proposed that a single antibody is made by separate gene segments that are brought together by recombination. According to this model, many copies of each gene segment are present, each slightly different from the others. Only one copy of each gene segment ends up in the final, recombined antibody gene (and hence in the antibody protein). Diversity is therefore achieved by combining different copies of each segment in different B cells as they mature.
43-11
At the time Dreyer and Bennett proposed their model, evidence for it was indirect. More than 10 years later, direct experimental evidence showed that their hypothesis is correct (Fig. 43.13). As a B cell differentiates, different gene segments are joined in a process called genomic rearrangement that produces a specific antibody.
FIG. 43.13: How is antibody diversity generated?
BACKGROUND Humans produce an estimated 10 billion different antibodies. Antibodies are proteins, encoded by genes. However, there are only about 25,000 genes in the human genome. How then can humans (and other vertebrates) produce such a diversity of antibodies from a limited set of genes? This was one of the central problems in immunology until the 1960s and 1970s.
HYPOTHESIS In 1965, American biologists William Dreyer and J. Claude Bennett proposed a model in which there are many copies of two gene segments, which they called the V (variable) and C (constant) segments. An antibody gene is assembled by recombining single copies of each of these segments. At the time, there was no direct experimental evidence to support this hypothesis.
EXPERIMENT In 1976, Japanese immunologists Nobumichi Hozumi and Susumu Tonegawa tested the Dreyer and Bennett hypothesis. Using mice as a model system, they isolated DNA from embryonic cells (before recombination was thought to occur) and from adult B cell tumor lines that produce one type of antibody (after recombination was thought to occur). The procedure they followed was slightly different from what is described here but conceptually the same. DNA is cut by restriction enzymes, the fragments are separated on a gel and transferred to a filter paper, and light chain DNA (containing both V and C regions) is used as a radioactive probe to determine the size and positions of the V and C regions (Southern blot, Chapter 12).
RESULTS Hozumi and Tonegawa found that the V and C regions were far apart in the DNA of embryonic cells and close together in adult cells, suggesting that the gene segments were brought together during B cell differentiation.
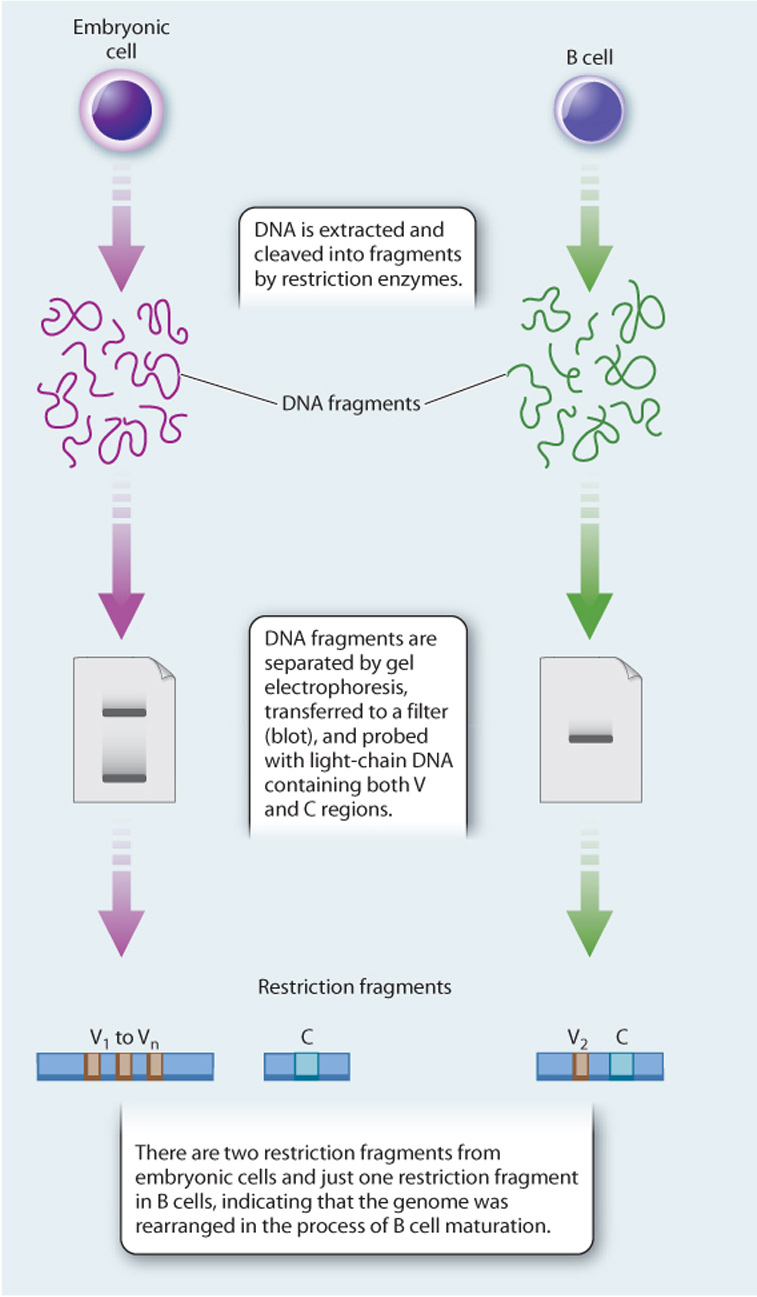
CONCLUSION Antibody genes are assembled by recombination of individual gene segments, providing a mechanism for generating antibody diversity.
FOLLOW-UP WORK Further studies showed that there are multiple copies of several different gene segments, now called the V, D (diversity), J (joining), and C regions for heavy chains, and V, J, and C regions for light chains. These are recombined in unique patterns in different B cells, so each B cell produces one antibody and a population of B cells produces a diversity of antibodies.
SOURCES Dreyer, W. J., and J. C. Bennett. 1965. “The Molecular Basis of Antibody Formation: A Paradox.” Proceedings of the National Academy of Sciences 54:864–869; Hozumi, N. and S. Tonegawa. 1976. “Evidence for Somatic Rearrangement of Immunoglobulin Genes Coding for Variable and Constant Regions.” Proceedings of the National Academy of Sciences 73:3628–3632.
43-12
For H chains, there are multiple V (variable), D (diversity), J (joining), and C (constant) gene segments (Fig. 43.14). In mice H chains, for example, there are 100 different V segments, 30 D segments, 6 J segments, and 8 C segments. During B cell differentiation, the DNA in this region undergoes recombination so that just one of each of these segments makes part of a functional H chain gene, and the intervening DNA is deleted. The assembled VDJ segment encodes the variable portion, and the C segment encodes the constant region. L chain genes also contain multiple copies of gene segments, except they do not have a D gene segment, so the VJ segment encodes the variable portion and the C segment encodes the constant region. The result of this process is that each B cell encodes a single H chain and a single L chain, and each B cell expresses a unique antibody.
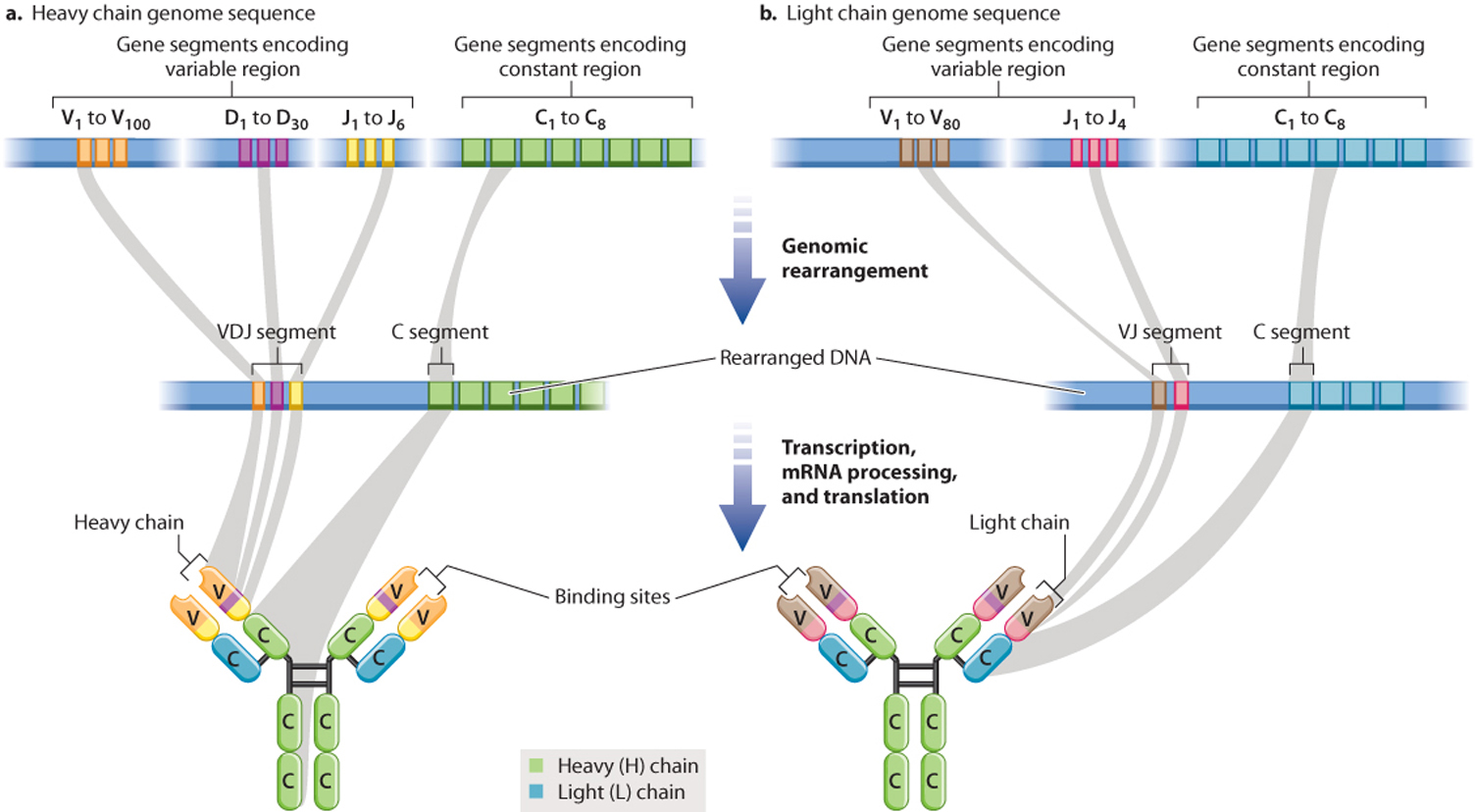
It is worth pausing here to consider the significance of this mechanism. We generally think of DNA as being a stable blueprint present in identical copies in all of the somatic cells in our bodies. B cells are an exception. As a result of genomic rearrangement, the DNA in each mature B cell is different from the DNA in every other mature B cell and different from the DNA in other cells in the body.
Genomic rearrangement is the key mechanism that creates antibody diversity, but other mechanisms contribute as well. One is simply the association of different L and H chains to make a functional antibody. In addition, recombination is sometimes imprecise, creating different sequences at the junctions between the different gene segments. There are also mechanisms for adding nucleotides at the junctions by a process called N‑nucleotide addition, and replacing nucleotides within VJ or VDJ segments by a process called somatic hypermutation. Finally, alternative splicing of mRNA (Chapter 3) allows a single B cell to express both membrane-bound and secreted antibodies.
We have seen that B cells as a group produce an enormous diversity of antibodies, whereas a given B cell makes one and only one antibody. But B cells are diploid, containing maternal and paternal copies of the genes that encode for antibodies. Maternal and paternal alleles of the antibody genes are similar, but not identical. As a result, a B cell able to use both homologs would make two different antibodies, yet B cells always make one. Remarkably, once one allele undergoes genomic rearrangement, the other allele is prevented from doing so. As a result, B cells act as if they have a single allele for each antibody gene—they express the maternal or the paternal gene, but not both.
43-13
As we have seen, B cells are able to switch between different classes, making IgM early in the response to infection and IgG later, while maintaining specificity to the same antigen. How is isotype switching achieved? Isotype switching results from further genome rearrangement, in which a given V(D)J region can join to different C segments during B cell differentiation.
The mechanisms we have considered pertain to humans and mice. Whereas B cells mature in the bone marrow in humans and mice, they mature in gut-associated lymphoid tissue (GALT) in many other vertebrates. Furthermore, genomic rearrangement is less important in many vertebrates. Instead, somatic hypermutation appears to play a more important role.