Chapter 11. Lab 11 Models in Biology II: Enzymatic Reactions
Learning Goals:
- Understand basic biochemical principles of proteins and enzymes
- Conduct an experiment with a physiologically important enzyme, acetylcholinesterase
- Learn how to use a standard curve to make indirect measurements
- Know the accuracies and limitations of models used in this lab
Lab Outline
Activity 1: Amino Acids and Protein Folding
Activity 2: Indirect Measurements of AChE
2A: Generating a Standard Curve
2B: Baseline Experiment
2C: Design Your Own Experiment
Scientific Inquiry
How do insects become resistant to pesticides? When a population of insects first comes into contact with a toxin (such as a pesticide), most members of that population die. But some survive...why? Have they acclimated to the pesticide? Did some of the insects not come into contact with the pesticide? The truth is much more interesting. As it turns out, some mutations are harmful, but occasionally, they are beneficial. In some organisms their genes have spontaneously changed, or mutated, naturally creating genetic diversity in a population. When a selection agent like a pesticide is introduced, those individual with beneficial mutations can resist pesticide activity and survive.
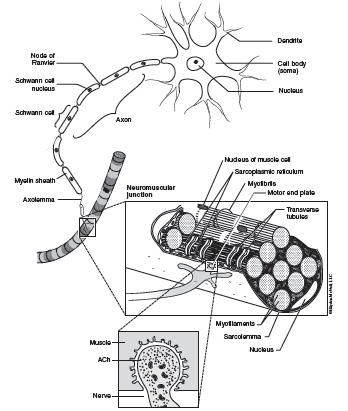
Scientists who study the insect survivors have discovered several beneficial mutations. Some insect larvae show increased amounts of proteins and lipids, and sclerotization of their protective cuticle that reduces the absorption of pesticides. Some resistant insects show an increase in the enzymes that metabolize the pesticides, while still others show a modification in the target protein’s active site so that the pesticide has no effect on the protein. To better understand this last difference, let’s examine a specific example. There is a large class of pesticides called organophosphates that target the protein acetylcholinesterase (AChE). If an insect lacks functional AChE, then it dies. What is the function of AChE? Let’s begin with how insects move. Insects have muscles that are quite similar to ours. When the insect needs to contract a muscle, a signal is sent from the nervous system, down a nerve, to the muscle. At the connection point between the nerve and muscle (the neuromuscular junction—Figure 1), the nerve releases a chemical called acetylcholine (ACh). This chemical travels across a very small space, called a synaptic cleft, and binds to receptors on the surface of the muscle cell which responds by contracting. AChE is an enzyme that is found in the synaptic cleft and catalyzes the hydrolysis of ACh into choline and acetate. AChE is a very important enzyme in insects because it is responsible for “stopping” the communication between the nerve and muscle cells. How do these pesticides inhibit the activity of AChE? Organophosphates bind to the active site of AChE and block the substrate (i.e. ACh) from binding. Some insects have evolved resistance to organophosphates through the mutation of one amino acid in the active site of AChE. This mutation still allows ACh to bind to the AChE, but it does not allow the organophosphate to bind to the AChE. In lab, ask your instructor to share the 3-D model of AChE to better understand this complex story.
Living cells contain a vast assortment of chemicals, such as ACh, which are necessary to sustain life. The interactions between these chemicals—the complex biochemical pathways through which material and energy are managed within a cell—are known collectively as metabolism. Metabolic interactions build up and break down molecules, and in the process store, use, or release energy. To understand these interactions, we first need to explore some basic biochemical principles. In this lab, we will explore these principles using various types of biological models. Models are important in biology because they allow us to visualize structures that we are unable to see with the naked eye. They also improve student learning by simplifying biological interactions. Today, you will have the chance to explore models of proteins and enzymes and then alter some environmental factors to examine the enzymatic activity of the actual AChE enzyme.
Background
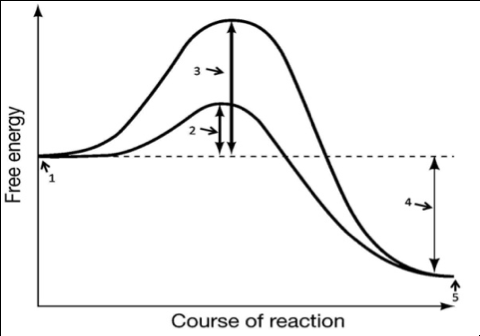
At the cellular level, numerous biochemical reactions are occurring within our bodies without our conscious awareness. Glucose, for example, is broken down by a complex series of controlled chemical reactions in order to generate adenosine triphosphate (ATP). In turn, ATP provides the cell with a readily available source of energy that can be used to drive other biochemical reactions. These reactions usually need extra energy to boost them over activation energy barriers. Heat can provide activation energy, but it is not a good option for living things since high heat denatures proteins (egg white protein turns from a clear viscous liquid to the solid denatured protein of cooked egg white, for example). Normal human body temperature (37 °C) is too low to activate most life-sustaining biochemical reactions so living systems have enzymes that lower biochemical activation energies making it possible for metabolic reactions to occur at normal body temperature. Without enzymes, metabolism would proceed very slowly at a time scale incompatible with human life as we know it. The following are important points to know about enzymes:
- Enzymes are biological catalysts that speed up biochemical reaction rates—they share the common feature of lowering activation energies required for metabolic reactions.
- By lowering activation energies, enzymes dramatically speed rates of chemical reactions by allowing far more molecules to participate in the reaction per unit time.
- Because enzymes are catalysts, they are not chemically altered by the reaction; rather, they form a transient complex with specific substrate molecules, and when the reaction is complete, the enzyme–product complex dissociates and the enzyme is free to catalyze another reaction.
- In addition, enzymes provide a high degree of specificity because they only interact with certain substrate molecules.
- Enzymes may differ with regard to the specific molecular mechanism by which they catalyze a reaction but they all work by chemically lowering the reaction’s activation energy.
Resources
Center for Biomolecular Modeling at the Milwaukee School of Engineering. 2008. Jmol Tutorials. http://cbm.msoe.edu/teachRes/jmol/ Accessed 2011 July.
Goodsell D. 2004. RCSB PDB Molecule of the Month. http://www.rcsb.org/pdb/home/home.do Accessed 2011 July.
Schomburg D. 2012. BRENDA: The comprehensive Enzyme Information System. http://www.brenda-enzymes.info/index.php4 Accessed 2 July 2013. Information on Acetylcholinesterase from Electrophorus electricus, such as optimal temperature and pH, is available in this database.
Schomburg I, Hofmann O, Baensch C, Chang A, Schomburg D. Enzyme data and metabolic information: BRENDA, a resource for research in biology, biochemistry, and medicine. Gene Funct. Dis. 2000;3–4:109–118.
Pre-Lab

Complete the BioPortal Quiz, which is designed to gauge your understanding of the prerequisites for this course, as well as your knowledge of the required content. It is your responsibility to review this material, if necessary, then watch the vodcast and read this lab. Place all notes in your lab notebook, which can be used during the Pre- Lab Quiz.
“Fold It” is a game used by research scientists to solve complex problems in structural biology. The program takes advantage of the variety of different skills and perspectives that each gamer brings to the problem. These multiple approaches provide unique ways of solving difficult problems. A gamer downloads the program and goes through a quick series of training exercises. Then they are given a proposed structure and are asked to resolve it to minimize its free energy. All of the physical and chemical rules are built into the boundaries of the game (so you don’t have to be a chemist or structural biologist to play). The element of competition drives gamers to use their individual creativity to get the best structures and earn a top score. The hope is that many collective minds working on the same problem will result in the rapid resolution of complex unknown structures. Link to game:http://fold.it/portal/info/science (This Software is loaded on all computers in the Lab and the BLC).
Print out the screen shot from your highest scoring structure and turn it in to your Instructor at the beginning of this lab to receive extra credit.
Activity 1: Amino Acids and Protein Folding
Learning Objectives
After successful completion of this activity, you should be able to:
- Demonstrate a salt bridge and disulfide bond using the amino acid kit
- Describe CPK coloring
- Distinguish hydrophobic and hydrophilic side chains
- Model the effect of pH on enzyme structure
Materials
Amino Acid Starter Kit
Computer with Swiss PDB Viewer
Procedure
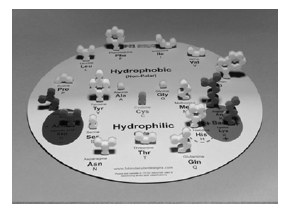
- First, look at the components in your Amino Acid Starter
Kit (Figure 2). Make sure that you have each of the
following:
- Chemical Properties Circle
- Amino Acid Chart
- Mini-Toober (foam-covered wire) Blue End Cap = N terminus; Red End Cap = C terminus
- 21 Foam Amino Acid Side Chain Models with Magnets
- 15 U-shaped Metal Clips
- Colors used in molecular models are important! Scientists
established the CPK Coloring Scheme to standardize identification
of specific atoms in models of molecular structure.
The Amino Acid Chart shows the atoms that compose
each side chain:
- Carbon is gray
- Oxygen is red
- Nitrogen is blue
- Hydrogen is white
- Sulfur is yellow
- Hydrophobic side chains are yellow
- Hydrophilic side chains are white
- Acidic side chains are red
- Basic side chains are blue
- Cysteine side chains are green
Hydrophobic side chains are composed primarily of __________ atoms.
Acidic side chains contain two __________ atoms. (This is a carboxylic acid functional group.)
Basic side chains contain __________ atoms. (This is an amino functional group.)
Hydrophilic side chains have various combinations of __________.
Is there an exception to any of these observations?
What atom do the metal clips on the toober represent?
Calculate the chance that your group’s amino acid chain is the same as the groups next to you. What does this tell you about protein diversity?
- Place each foam amino acid side chain on the magnetic Chemical Properties Circle according to its chemical properties. If necessary, consult the Amino Acid Side Chain List. Turn the gray sides towards you. What are some commonalities among the hydrophobic side chains? What about the hydrophilic ones?
- Unwind the 4-foot mini-toober (foam covered wire) in your kit. Notice the blue and red end caps on the ends of your mini-toober. The blue end cap represents the N-terminus (the beginning) of the protein, and the red end cap represents the C-terminus (the end) of the protein.
- Select 15 metal u-shaped clips from your kit. Beginning at the N-terminus of your mini-toober, measure about three inches from the end of your mini-toober and slide the first clip into place. Place the rest of the clips three inches apart on your mini-toober until all are attached to the mini-toober.
- Choose 15 side chains from the Chemical Properties Circle and place them (in any order you choose) on your mini-toober. Use this list for your selection:
- 6 hydrophobic side chains
- 2 acidic side chains
- 2 basic side chains
- 2 cysteine side chains
What would the effects of a pH change be on the protein you built?
What kind of mutations do you think would have the most damaging effects on the protein? What kinds of mutations would have a lesser effect?
- Record the 15 amino acid sequence of your protein in your lab notebook.
- Fold it! Follow these basic principles to bring your amino acid to a low energy state.
- Remember that your protein is folding in water. Start by folding your protein so that all of the hydrophobic side chains are buried on the inside of your protein.
- Next, fold your protein so the acidic and basic (charged) side chains are on the outside surface of the protein and pair one negative side chain with one positive side chain so that they come within one inch of each other and neutralize each other. This positive-negative pairing is called a “salt bridge” and helps stabilize your protein.
- Continue to fold your protein making sure that the polar side chains are on the outside surface of your protein where they can hydrogen bond with water.
- Finally, fold your protein so that the two cysteine side chains are positioned opposite each other on the inside of the protein where they can form a covalent disulfide bond that helps stabilize your protein.
- Note: As you continue to fold your protein to apply each new property listed above, you will probably find that some of the side chains you previously positioned are no longer in place. Continue to fold until the hydrophobic ones are buried on the inside again. Find a shape in which all the properties apply.
- The final shape of your protein when it is folded is called the Tertiary Structure. Sketch the structure of your protein in your lab notebook or take a picture of it. Put your model aside for now, we will use it later in lab.
- Your Instructor will guide you through the use of your model to demonstrate concepts such as active site and protein denaturation.
- Think of the strengths and weaknesses of this type of model and record that in the table in your lab notebook.
Activity 2: Indirect Measurement of Enzymatic Activity of Acetylcholinesterase
In today’s lab you will have an opportunity to monitor the activity of acetylcholinesterase from Electrophorus electricus (electric eel). This enzyme is the target of many pesticides that are sprayed on crops to rid them of insect pests. Acetylcholinesterase (AChE) hydrolyses the neuro-transmitter acetylcholine (ACh) into acetic acid and choline. The normal reaction for AChE is shown below. Based on this knowledge, how would you monitor the activity of AChE? If you had a test tube containing the substrate acetylcholine and the enzyme acetylcholinesterase, you might measure the decline in substrate (ACh) or increase in products (acetic acid or choline) over time as a result of enzyme activity. How would you monitor these changes?
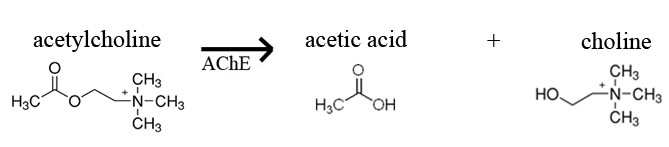
In this lab, we will use commercially produced acetylthiocholine (ATCh) as the substrate for acetylcholinesterase activity instead of the natural substrate, acetylcholine (ACh). AChE hydrolyzes ATCh into acetic acid and thiocholine (see reaction below).
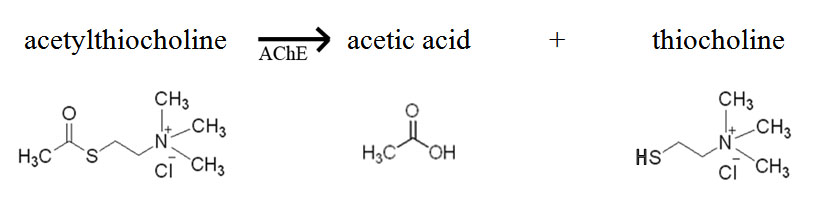
Why are we using a modified substrate rather than the actual substrate to study the activity of this enzyme? What are some possible drawbacks to using a substitute substrate?
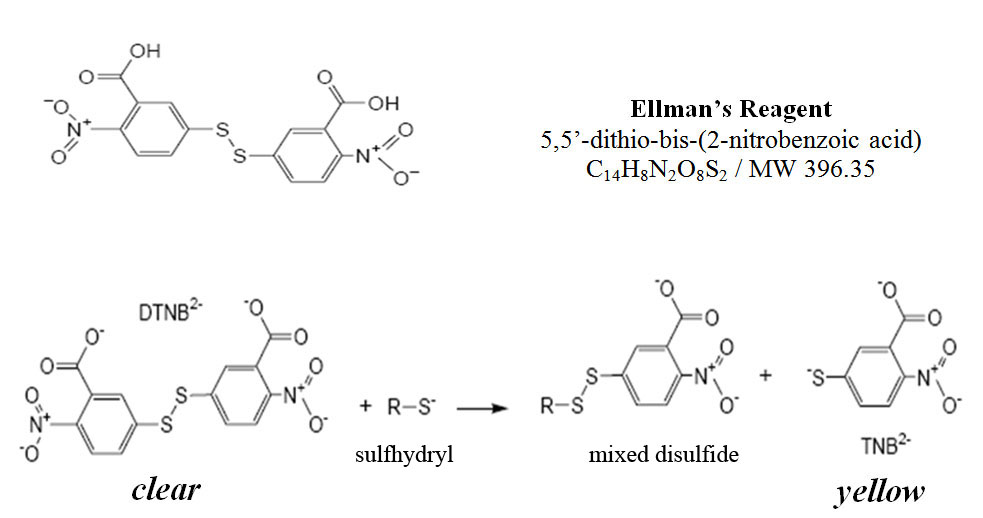
Thiocholine, one of the products of ATCh hydrolysis by AChE, is called a thiol because it is an organic molecule with a sulfhydryl group (–SH). This free sulfhydryl group can be detected by using the compound DTNB, known as Ellman’s reagent. DTNB is clear in the absence of sulfhydryl groups, but is reduced by free sulfhydryl groups to produce a yellow byproduct (TNB2–). The reaction for reduction of Ellman’s reagent is shown below. The intensity of the yellow color varies with the concentration of sulfhydryl present; therefore the darkness, or intensity of the yellow color produced allows us to indirectly measure the concentration of thiocholine present using a spectrophotometer. TNB2– gives maximal absorption at a wavelength of 412 nm (Pierce Biotechnology, Inc. 2004).
DTNB reacts with sulfhydryl containing compounds, represented here by R-S, to produce the yellow TNB2–.
Converting from Absorbance to Molarity: Using a Standard Curve
During the course of the enzyme reaction, you will use a spectrophotometer to record the absorbance of the solution as it changes color from clear to yellow. Recall that acetylcholinesterase (AChE) hydrolyzes acetylthiocholine into acetic acid and thiocholine. Thiocholine reacts with DTNB to produce TNB2–, which is yellow in color. How can absorbance units be converted into a molar concentration of thiocholine?
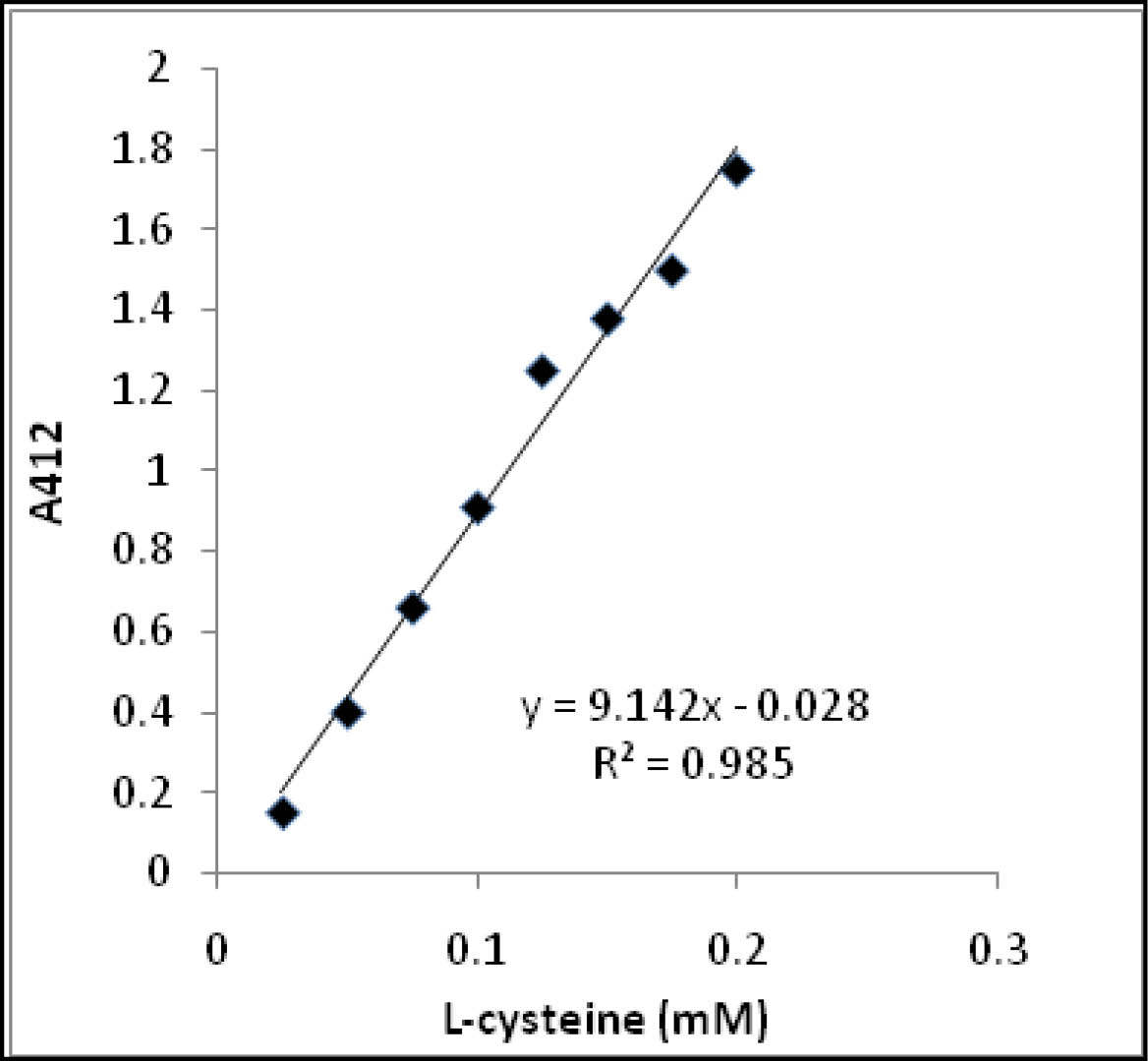
You can create a standard curve to compare the amount of TNB2– in solution to absorbance in the spectrophotometer. You can do this by preparing different concentrations of TNB2– and measuring their corresponding absorbances. Then, plot the concentration and absorbance to produce a correlation line relating known concentrations of TNB2– to measured absorbance (Figure 3). This is called a standard curve and it shows the relationship between the concentration of a substance and its absorbance at a particular wavelength of light (in this case, 412 nm).
Houston, we have a problem…
What if we don’t have TNB2–? What else could we use? Do you know of any chemical that has a free sulfhydryl group? What about the amino acid L-cysteine? It has one sulfhydryl group just like thiocholine. For this lab, you will use L-cysteine to create your standard curve (see Figure 3).
What would you expect the reaction of DTNB with L-cysteine to look like?
Learning Objectives
After successful completion of this activity, you should be able to:
- Use the Spectrophotometer to determine the absorbance of a solution
- Perform an experiment to test the rate of reaction of an enzyme
- Use a standard curve to convert from absorbance to molarity
Materials
7.5 mM Acetylthiocholine (ATCh) made in PB7
5 mM 5,5’-dithio-bis (2-nitrobenzoic acid) = DTNB, made in PB7 containing 0.1mM EDTA
5 μg/ml AChE prepared in PB7 with BSA; enzyme isolated from E. electricus (Sigma)
Spectrophotometer
pH meter
Vortex
p100 and p1000
Pipette tips
Test tubes
3 ml transfer pipettes
Ice
Activity 2A: Generating a Standard Curve
Procedure
- As a section, you will receive 6 blanks and 6 standard concentration solutions of 1 mM cysteine in varying amounts of DTNB. The final concentrations of cysteine in these tubes range from 0.025 mM to 0.2 mM. Each lab group should take 1 blank tube and one of the standards.
- Blank the Spec20 using the tube that contains buffer and DTNB, without cysteine. Refer to Appendix B if you are unable to remember how to do this from Lab 2.
- Measure the A412 of the standard in the spectrophotometer.
- Record the concentration of cysteine and its corresponding absorbance as a section.
- Graph your results using Microsoft Excel™ (see sample output in Figure 3). Insert a trendline and display the equation of a line along with the R2 value.
Does a linear trendline fit your data or should you use a curve? Explain your answer.
- Read your Standard Curve. Notice that in the sample curve (Figure 11.6), the concentration of 0.05 mM corresponds to an A412 value of 0.4. Since the standard curve is a continuous line, you can read the concentration of your solution by interpolation of any absorbance reading as long as the readings lie within your data range. Use your section’s standard curve as well as the absorbance readings of your experimental tubes (in Activity 2B) to determine their thiocholine concentration.
Based on the sample standard curve in Figure 3, if you read A412 = 1.0 for your AChE reaction, what would be the concentration of thiocholine in the tube?
Activity 2B: Baseline Experiment
Measure the rate of acetylcholinesterase activity at room temperature in the standard buffer PB7 (0.1 M sodium phosphate buffer, pH 7).
- Label two tubes, one “E” for experimental and one “C” for control. The directions for the assay are given in steps below as well as summarized in Table 1.
- Add 3.5 ml of PB7 to the experimental tube and 3.6 ml of PB7 to the control tube. Keep track of the reagents you have added by checking off each reagent as it is added to the tube.
- Add 200 μl of 7.5 mM ATCh (substrate) to the control test tube. Vortex gently to mix. Repeat for the experimental tube.
- Add 200 μl of 5 mM DTNB to the control test tube. Vortex gently to mix. Repeat for the experimental tube.
- Use the control tube to blank the Spec20. Why?
Is this a positive or negative control?
- STOP! Read ahead to be prepared for the next steps before starting your reaction. When you are ready, add 100 μl of AChE (enzyme) to the experimental tube and vortex gently to mix. Start your timer.
- Quickly place the experimental tube in the Spec20 and leave it there. Immediately take an initial reading.
- Record the A412 values every 30 seconds for a total of 5 minutes (11 readings).
- At the end of the 5 minutes, read the control tube again and record its A412 value. Has it changed? If so, why? What will you do if it is different from its starting value?
The enzyme has been prepared in sodium phosphate buffer.
An additional 0.1 ml of PB7 was included in the control in lieu of the enzyme.
Table 1 Summary of reagents for control and experimental tubes.
Data Analysis
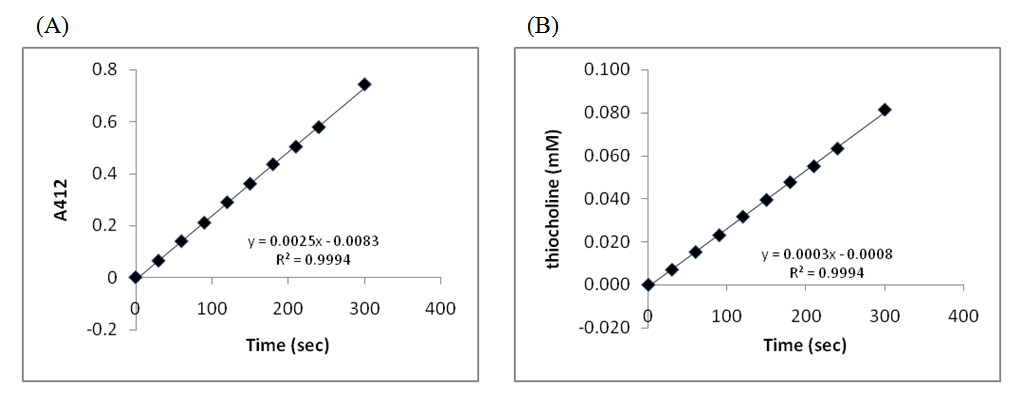
- Graph your results (Figure 4A is a sample).
- Based on the A412 values, determine the amount of product generated in each tube for each time point using the standard curve you generated in Lab Activity 2A (Figure 4B is a sample).
- Determine the rate of the reaction for the Standard Protocol. Did the rate of the reaction change during the course of the experiment? Why or why not?
- Sketch the curves for the change in (1) substrate concentration over time; (2) thiocholine concentration over time; and (3) AChE concentration over time.
Activity 2C: Design Your Own Experiment
In this activity, you will perform an additional AChE experiment of your own design. Of the following two conditions, each group will choose two temperatures or two pH values.
Effects of Temperature on AChE Activity
- Select two of the following temperatures: 0°C, 30°C, 37°C or 50°C.
- Prepare your reaction tubes (without enzyme) so that you can monitor one temperature at a time.
- Use the most appropriate method to implement your experiment and collect your data. There are at least two basic experimental strategies for testing temperature effects on AChE:
a. Incubate the AChE enzyme at various temperatures (e.g., ice, 30 or 50) for an amount of time that you choose and then add the heat-treated enzyme to the reaction tube at room temperature
b. Pre-warm the reaction mixture without enzyme to the desired temperatures and then add AChE and measure A412 at given time points. In this scenario, you will return the test tubes to the heating blocks between readings.
What is the difference between these two approaches to temperature?
Which should you use and why?
Here are some helpful hints:
- It requires at least 10 minutes for the reaction mixture to reach 50°C. Will you pre-warm your tubes?
- Will you preheat the buffer or will you also include the other reagents, excluding the enzyme?
- Heating may cause changes to the reaction mixture. How will you compensate for this?
Effects of [H+] on AChE Activity
- Select two of the following options for [H+]: pH 5, 6, 8, or 9.
- Prepare your reaction tubes (without enzyme) so that you can monitor one pH at a time.
- Implement your experiment and collect your data.
Here are some helpful hints:
- Substitute PB7 with PB 5, 6, 8, or 9.
- How will you verify the pH of your reaction mixtures? Should you read the pH of the mixtures at the end of the reaction using a pH meter?
- Particular pH values may cause changes to the reaction mixture. How will you deal with this?
Sample Design Procedure
- If you are testing the effect of pH 5 on AChE activity, make a control tube and experimental tube each containing pH 5.0 buffer.
- Add the ATCh and DTNB to each tube, as before.
- Blank the Spec20 with the control tube and place the tube to the side. Add the 100 μl of AChE to the experimental tube and begin recording data.
- Repeat this procedure for the other pH value that you wish to test.
- Use the standard curve to determine the concentration of thiocholine at each time point, and graph the concentration of thiocholine versus time (Figure 4B).
- Determine the rate of the reaction for each pH tested. What type of figure will you use for this? What conclusions can you draw?
Class Discussion

You will present the data from your “Design your Own” AChE experiment at the end of lab. You should prepare to present the following information.
- Describe the nature of your experiments, the hypotheses tested and the predicted outcomes.
- Identify the variables (enzyme, substrate, pH, and temperature) of your experiments.
- Include tables and graphs to describe your results. Explain each carefully.
- Interpret and explain what you think happened to yield your results.
- State whether your results support your original hypotheses and explain your reasoning.
- Briefly discuss your results. What questions were raised in the course of your investigations?
Self-Assessment
These questions were taken from previous exams and are meant to represent a sample, not a complete study guide. The questions in these examples are designed to test your understanding of the concepts and skills presented in this lab, and your ability to apply what you have learned to novel problems.
Question 11.1
Computation: Determine the slope equation (y = mx + b, where the intercept b is set to “0”) for “y vs. x” using the values in columns 1 and 2 of the table. Use this equation to calculate the unknown “x” in column 4 for a given “y” in column 3.”
| 1 | 2 | 3 | 4 |
|---|---|---|---|
| x | y | y | x |
| 1 | 2 | 2.5 | DYU2tVvtzEQ= |
| 2 | 4 | 4.8 | 5OE3htGCBq8= |
| 3 | 6 | 7 | a+gNtjpaMI8= |
| 4 | 8 | 10.4 | LRu7BWCWudE= |