PROBLEMS
WORKING WITH THE FIGURES
Question 4.1
In Figure 4-3, would there be any meiotic products that did not undergo a crossover in the meiosis illustrated? If so, what colors would they be in the color convention used?
Question 4.2
In Figure 4-6, why does the diagram not show meioses in which two crossovers occur between the same two chromatids (such as the two inner ones)?
Question 4.3
In Figure 4-8, some meiotic products are labeled parental. Which parent is being referred to in this terminology?
Question 4.4
In Figure 4-9, why is only locus A shown in a constant position?
Question 4.5
In Figure 4-10, what is the mean frequency of crossovers per meiosis in the region A-
Question 4.6
In Figure 4-11, is it true to say that from such a cross the product v cv+ can have two different origins?
Question 4.7
In Figure 4-14, in the bottom row four colors are labeled SCO. Why are they not all the same size (frequency)?
Question 4.8
Using the conventions of Figure 4-15, draw parents and progeny classes from a cross
P M′″/p M′ × p M′/p M″″
Question 4.9
In Figure 4-17, draw the arrangements of alleles in an octad from a similar meiosis in which the upper product of the first division segregated in an upside-
Question 4.10
In Figure 4-19, what would be the RF between A/a and B/b in a cross in which purely by chance all meioses had four-
Question 4.11
In Figure 4-21, let GC = A and AT = a, then draw the fungal octad that would result from the final structure (5).
(Challenging) Insert some closely linked flanking markers into the diagram, say P/p to the left and Q/q to the right (assume either cis or trans arrangements). Assume neither of these loci show non-
Mendelian segregation. Then draw the final octad based on the structure in part 5.
162
BASIC PROBLEMS
Question 4.12
A plant of genotype
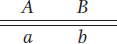
is testcrossed with
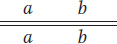
If the two loci are 10 m.u. apart, what proportion of progeny will be AB/ab?
Question 4.13
The A locus and the D locus are so tightly linked that no recombination is ever observed between them. If Ad/Ad is crossed with aD/aD and the F1 is intercrossed, what phenotypes will be seen in the F2 and in what proportions?
Question 4.14
The R and S loci are 35 m.u. apart. If a plant of genotype
is selfed, what progeny phenotypes will be seen and in what proportions?
Question 4.15
The cross E/E · F/F × e/e · f/f is made, and the F1 is then backcrossed with the recessive parent. The progeny genotypes are inferred from the phenotypes. The progeny genotypes, written as the gametic contributions of the heterozygous parent, are in the following proportions:
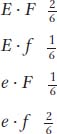
Explain these results.
Question 4.16
A strain of Neurospora with the genotype H · I is crossed with a strain with the genotype h · i. Half the progeny are H · I, and the other half are h · i. Explain how this outcome is possible.
Question 4.17
A female animal with genotype A/a · B/b is crossed with a double-
Question 4.18
If A/A · B/B is crossed with a/a · b/b and the F1 is testcrossed, what percentage of the testcross progeny will be a/a · b/b if the two genes are (a) unlinked; (b) completely linked (no crossing over at all); (c) 10 m.u. apart; (d) 24 m.u. apart?
Question 4.19
In a haploid organism, the C and D loci are 8 m.u. apart. From a cross C d × c D, give the proportion of each of the following progeny classes: (a) C D; (b) c d; (c) C d; (d) all recombinants combined.
Question 4.20
A fruit fly of genotype B R/b r is testcrossed with b r/b r. In 84 percent of the meioses, there are no chiasmata between the linked genes; in 16 percent of the meioses, there is one chiasma between the genes. What proportion of the progeny will be B r/b r?
Question 4.21
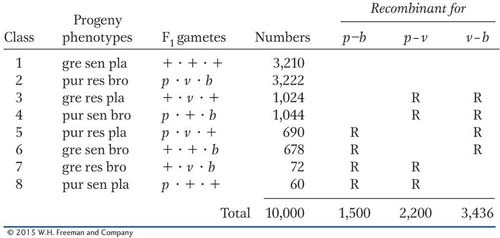
A three-
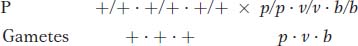
Determine which genes are linked.
Draw a map that shows distances in map units.
Calculate interference, if appropriate.
163
 Unpacking Problem 21
Unpacking Problem 21
Sketch cartoon drawings of the P, F1, and tester corn plants, and use arrows to show exactly how you would perform this experiment. Show where seeds are obtained.
Why do all the +’s look the same, even for different genes? Why does this not cause confusion?
How can a phenotype be purple and brown, for example, at the same time?
Is it significant that the genes are written in the order p-
v- b in the problem? What is a tester and why is it used in this analysis?
What does the column marked “Progeny phenotypes” represent? In class 1, for example, state exactly what “gre sen pla” means.
What does the line marked “Gametes” represent, and how is it different from the column marked “F1 gametes”? In what way is comparison of these two types of gametes relevant to recombination?
Which meiosis is the main focus of study? Label it on your drawing.
Why are the gametes from the tester not shown?
Why are there only eight phenotypic classes? Are there any classes missing?
What classes (and in what proportions) would be expected if all the genes are on separate chromosomes?
To what do the four pairs of class sizes (very big, two intermediates, very small) correspond?
What can you tell about gene order simply by inspecting the phenotypic classes and their frequencies?
What will be the expected phenotypic class distribution if only two genes are linked?
What does the word “point” refer to in a three-
point testcross? Does this word usage imply linkage? What would a four- point testcross be like? What is the definition of recombinant, and how is it applied here?
What do the “Recombinant for” columns mean?
Why are there only three “Recombinant for” columns?
What do the R’s mean, and how are they determined?
What do the column totals signify? How are they used?
What is the diagnostic test for linkage?
What is a map unit? Is it the same as a centimorgan?
In a three-
point testcross such as this one, why aren’t the F1 and the tester considered to be parental in calculating recombination? (They are parents in one sense.) What is the formula for interference? How are the “expected” frequencies calculated in the coefficient-
of- coincidence formula? Why does part c of the problem say “if appropriate”?
How much work is it to obtain such a large progeny size in corn? Which of the three genes would take the most work to score? Approximately how many progeny are represented by one corncob?
Question 4.22
You have a Drosophila line that is homozygous for autosomal recessive alleles a, b, and c, linked in that order. You cross females of this line with males homozygous for the corresponding wild-
What is the recombinant frequency between a and b? Between b and c? (Remember, there is no crossing over in Drosophila males.)
What is the coefficient of coincidence?
Question 4.23
R. A. Emerson crossed two different pure-
What were the genotypes of the parental lines?
Draw a linkage map for the three genes (include map distances).
Calculate the interference value.
Question 4.24
Chromosome 3 of corn carries three loci (b for plant-
Question 4.25
Groodies are useful (but fictional) haploid organisms that are pure genetic tools. A wild-
164
Question 4.26
In Drosophila, the allele dp+ determines long wings and dp determines short (“dumpy”) wings. At a separate locus, e+ determines gray body and e determines ebony body. Both loci are autosomal. The following crosses were made, starting with pure-
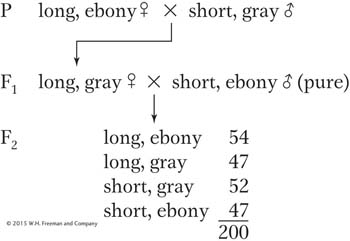
Use the χ2 test to determine if these loci are linked. In doing so, indicate (a) the hypothesis, (b) calculation of χ2, (c) p value, (d) what the p value means, (e) your conclusion, (f) the inferred chromosomal constitutions of parents, F1, tester, and progeny.
Question 4.27
The mother of a family with 10 children has blood type Rh+. She also has a very rare condition (elliptocytosis, phenotype E) that causes red blood cells to be oval rather than round in shape but that produces no adverse clinical effects. The father is Rh− (lacks the Rh+ antigen) and has normal red blood cells (phenotype e). The children are 1 Rh+ e, 4 Rh+ E, and 5 Rh− e. Information is available on the mother’s parents, who are Rh+ E and Rh− e. One of the 10 children (who is Rh+ E) marries someone who is Rh+ e, and they have an Rh+ E child.
Draw the pedigree of this whole family.
Is the pedigree in agreement with the hypothesis that the Rh+ allele is dominant and Rh− is recessive?
What is the mechanism of transmission of elliptocytosis?
Could the genes governing the E and Rh phenotypes be on the same chromosome? If so, estimate the map distance between them, and comment on your result.
Question 4.28
From several crosses of the general type A/A · B/B × a/a · b/b, the F1 individuals of type A/a · B/b were testcrossed with a/a · b/b. The results are as follows:
|
Testcross of F1 from cross |
Testcross progeny |
|||
|---|---|---|---|---|
|
A/a · B/b |
a/a · b/b |
A/a · b/b |
a/a · B/b |
|
|
1 |
310 |
315 |
287 |
288 |
|
2 |
36 |
38 |
23 |
23 |
|
3 |
360 |
380 |
230 |
230 |
|
4 |
74 |
72 |
50 |
44 |
For each set of progeny, use the χ2 test to decide if there is evidence of linkage.
Question 4.29
In the two pedigrees diagrammed here, a vertical bar in a symbol stands for steroid sulfatase deficiency, and a horizontal bar stands for ornithine transcarbamylase deficiency.
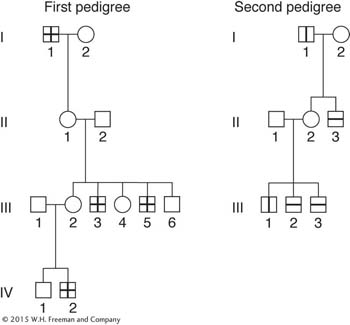
Is there any evidence in these pedigrees that the genes determining the deficiencies are linked?
If the genes are linked, is there any evidence in the pedigree of crossing over between them?
Assign genotypes of these individuals as far as possible.
Question 4.30
In the accompanying pedigree, the vertical lines stand for protan color blindness, and the horizontal lines stand for deutan color blindness. These are separate conditions causing different misperceptions of colors; each is determined by a separate gene.
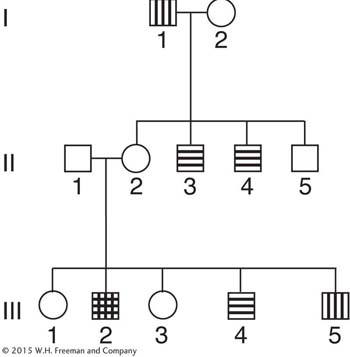
165
Does the pedigree show any evidence that the genes are linked?
If there is linkage, does the pedigree show any evidence of crossing over?
Explain your answers to parts a and b with the aid of the diagram.
Can you calculate a value for the recombination between these genes? Is this recombination by independent assortment or by crossing over?
Question 4.31
In corn, a triple heterozygote was obtained carrying the mutant alleles s (shrunken), w (white aleurone), and y (waxy endosperm), all paired with their normal wild-
Determine if any of these three loci are linked and, if so, show map distances.
Show the allele arrangement on the chromosomes of the triple heterozygote used in the testcross.
Calculate interference, if appropriate.
Question 4.32
A mouse cross A/a · B/b χ a/a · b/b is made, and in the progeny there are
25% A/a • B/b, 25% a/a • b/b,
25% A/a • b/b, 25% a/a • B/b
Explain these proportions with the aid of simplified meiosis diagrams.
A mouse cross C/c · D/d × c/c · d/d is made, and in the progeny there are
45% C/c • d/d, 45% c/c • D/d,
5% c/c • d/d, 5% C/c • D/d
Explain these proportions with the aid of simplified meiosis diagrams.
Question 4.33
In the tiny model plant Arabidopsis, the recessive allele hyg confers seed resistance to the drug hygromycin, and her, a recessive allele of a different gene, confers seed resistance to herbicide. A plant that was homozygous hyg/hyg · her/her was crossed with wild type, and the F1 was selfed. Seeds resulting from the F1 self were placed on petri dishes containing hygromycin and herbicide.
If the two genes are unlinked, what percentage of seeds are expected to grow?
In fact, 13 percent of the seeds grew. Does this percentage support the hypothesis of no linkage? Explain. If not, calculate the number of map units between the loci.
Under your hypothesis, if the F1 is testcrossed, what proportion of seeds will grow on the medium containing hygromycin and herbicide?
Question 4.34
In a diploid organism of genotype A/a; B/b; D/d, the allele pairs are all on different chromosome pairs. The two diagrams below purport to show anaphases (“pulling apart” stages) in individual cells. State whether each drawing represents mitosis, meiosis I, or meiosis II or is impossible for this particular genotype.
Question 4.35
The Neurospora cross al-2+ × al-2 is made. A linear tetrad analysis reveals that the second-
Draw two examples of second-
division segregation patterns in this cross. What can be calculated by using the 8 percent value?
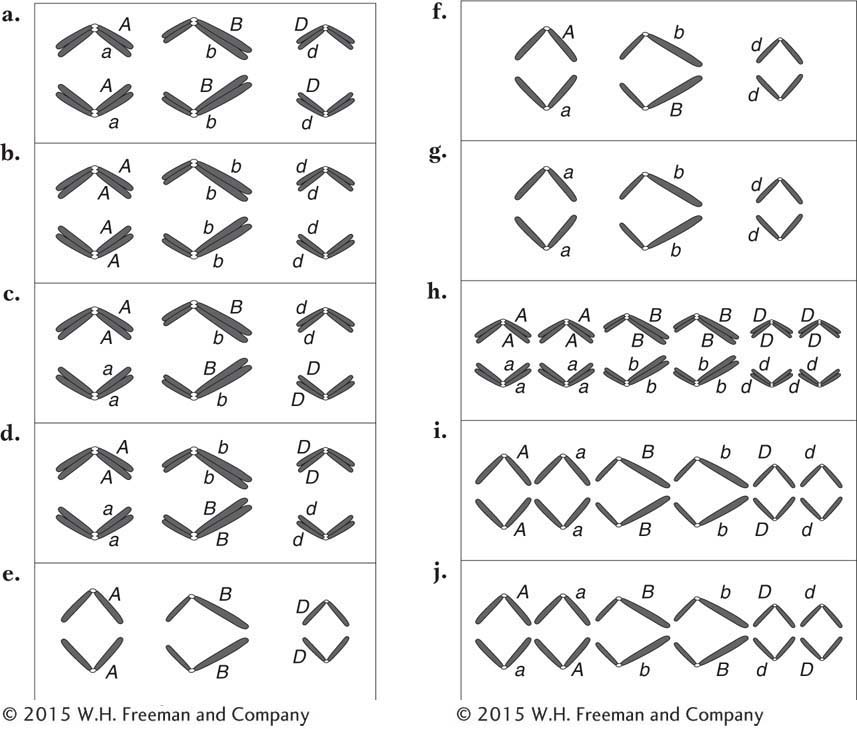
166
Question 4.36
From the fungal cross arg-
Question 4.37
For a certain chromosomal region, the mean number of crossovers at meiosis is calculated to be two per meiosis. In that region, what proportion of meioses are predicted to have (a) no crossovers? (b) one crossover? (c) two crossovers?
Question 4.38
A Neurospora cross was made between a strain that carried the mating-
Deduce the linkage arrangement of the mating-
type locus and the arg- 1 locus. Include the centromere or centromeres on any map that you draw. Label all intervals in map units. Diagram the meiotic divisions that led to class 6. Label clearly.
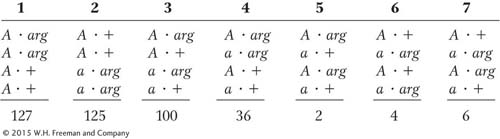
 Unpacking Problem 38
Unpacking Problem 38
Are fungi generally haploid or diploid?
How many ascospores are in the ascus of Neurospora? Does your answer match the number presented in this problem? Explain any discrepancy.
What is mating type in fungi? How do you think it is determined experimentally?
Do the symbols A and a have anything to do with dominance and recessiveness?
What does the symbol arg-
1 mean? How would you test for this genotype? How does the arg-
1 symbol relate to the symbol +? What does the expression wild type mean?
What does the word mutant mean?
Does the biological function of the alleles shown have anything to do with the solution of this problem?
What does the expression linear octad analysis mean?
In general, what more can be learned from linear tetrad analysis that cannot be learned from unordered tetrad analysis?
How is a cross made in a fungus such as Neurospora? Explain how to isolate asci and individual ascospores. How does the term tetrad relate to the terms ascus and octad?
Where does meiosis take place in the Neurospora life cycle? (Show it on a diagram of the life cycle.)
What does Problem 38 have to do with meiosis?
Can you write out the genotypes of the two parental strains?
Why are only four genotypes shown in each class?
Why are there only seven classes? How many ways have you learned for classifying tetrads generally? Which of these classifications can be applied to both linear and unordered tetrads? Can you apply these classifications to the tetrads in this problem? (Classify each class in as many ways as possible.) Can you think of more possibilities in this cross? If so, why are they not shown?
Do you think there are several different spore orders within each class? Why would these different spore orders not change the class?
Why is the following class not listed?
a · +
A · arg
a · +
A · arg
What does the expression linkage arrangement mean?
What is a genetic interval?
Why does the problem state “centromere or centromeres” and not just “centromere”? What is the general method for mapping centromeres in tetrad analysis?
What is the total frequency of A · + ascospores? (Did you calculate this frequency by using a formula or by inspection? Is this a recombinant genotype? If so, is it the only recombinant genotype?)
The first two classes are the most common and are approximately equal in frequency. What does this information tell you? What is their content of parental and recombinant genotypes?
Question 4.39
A geneticist studies 11 different pairs of Neurospora loci by making crosses of the type a · b × a+ · b+ and then analyzing 100 linear asci from each cross. For the convenience of making a table, the geneticist organizes the data as if all 11 pairs of genes had the same designation—
167
|
Number of asci of type |
|||||||
|---|---|---|---|---|---|---|---|
|
Cross |
a · b |
a · b+ |
a · b |
a · b |
a · b |
a · b+ |
a · b+ |
|
a · b |
a · b+ |
a · b+ |
a+ · b |
a+ · b+ |
a+ · b |
a+ · b |
|
|
a+ · b+ |
a+ · b |
a+ · b+ |
a+ · b+ |
a+ · b+ |
a+ · b |
a+ · b+ |
|
|
a+ · b+ |
a+ · b |
a+ · b |
a · b+ |
a · b |
a · b+ |
a · b |
|
|
1 |
34 |
34 |
32 |
0 |
0 |
0 |
0 |
|
2 |
84 |
1 |
15 |
0 |
0 |
0 |
0 |
|
3 |
55 |
3 |
40 |
0 |
2 |
0 |
0 |
|
4 |
71 |
1 |
18 |
1 |
8 |
0 |
1 |
|
5 |
9 |
6 |
24 |
22 |
8 |
10 |
20 |
|
6 |
31 |
0 |
1 |
3 |
61 |
0 |
4 |
|
7 |
95 |
0 |
3 |
2 |
0 |
0 |
0 |
|
8 |
6 |
7 |
20 |
22 |
12 |
11 |
22 |
|
9 |
69 |
0 |
10 |
18 |
0 |
1 |
2 |
|
10 |
16 |
14 |
2 |
60 |
1 |
2 |
5 |
|
11 |
51 |
49 |
0 |
0 |
0 |
0 |
0 |
Question 4.40
Three different crosses in Neurospora are analyzed on the basis of unordered tetrads. Each cross combines a different pair of linked genes. The results are shown in the following table:
|
Cross |
Parents (%) |
Parental ditypes (%) |
Tetratypes (%) |
Nonparental ditypes (%) |
|---|---|---|---|---|
|
1 |
a · b+ × a+ · b |
51 |
45 |
4 |
|
2 |
c · d+ × c+ · d |
64 |
34 |
2 |
|
3 |
e · f+ × e+ · f |
45 |
50 |
5 |
For each cross, calculate
the frequency of recombinants (RF).
the uncorrected map distance, based on RF.
the corrected map distance, based on tetrad frequencies.
the corrected map distance, based on the mapping function.
Question 4.41
On Neurospora chromosome 4, the leu3 gene is just to the left of the centromere and always segregates at the first division, whereas the cys2 gene is to the right of the centromere and shows a second-
|
(i) l c |
(ii) l + |
(iii) l c |
(iv) l c |
(v) l c |
(vi) l + |
(vii) l + |
|
l c |
l + |
l + |
+ c |
+ + |
+ c |
+ c |
|
+ + |
+ c |
+ + |
+ + |
+ + |
+ c |
+ + |
|
+ + |
+ c |
+ c |
l + |
l c |
l + |
l c |
Question 4.42
A rice breeder obtained a triple heterozygote carrying the three recessive alleles for albino flowers (al), brown awns (b), and fuzzy leaves (fu), all paired with their normal wild-
|
170 |
wild type |
|
150 |
albino, brown, fuzzy |
|
5 |
brown |
|
3 |
albino, fuzzy |
|
710 |
albino |
|
698 |
brown, fuzzy |
|
42 |
fuzzy |
|
38 |
albino, brown |
Are any of the genes linked? If so, draw a map labeled with map distances. (Don’t bother with a correction for multiple crossovers.)
The triple heterozygote was originally made by crossing two pure lines. What were their genotypes?
Question 4.43
In a fungus, a proline mutant (pro) was crossed with a histidine mutant (his). A nonlinear tetrad analysis gave the following results:
|
+ |
+ |
+ |
+ |
+ |
his |
|
+ |
+ |
+ |
his |
+ |
his |
|
pro |
his |
pro |
+ |
pro |
+ |
|
pro |
his |
pro |
his |
pro |
+ |
|
6 |
82 |
112 |
|||
Are the genes linked or not?
Draw a map (if linked) or two maps (if not linked), showing map distances based on straightforward recombinant frequency where appropriate.
If there is linkage, correct the map distances for multiple crossovers (choose one approach only).
Question 4.44
In the fungus Neurospora, a strain that is auxotrophic for thiamine (mutant allele t) was crossed with a strain that is auxotrophic for methionine (mutant allele m). Linear asci were isolated and classified into the following groups:
168
|
Spore pair |
Ascus types |
|||||
|---|---|---|---|---|---|---|
|
1 and 2 |
t+ |
t+ |
t+ |
t+ |
t m |
t m |
|
3 and 4 |
t+ |
t m |
+ m |
++ |
t m |
++ |
|
5 and 6 |
+ m |
++ |
t+ |
t m |
++ |
t+ |
|
7 and 8 |
+ m |
+ m |
+ m |
+ m |
+ + |
+ m |
|
Number |
260 |
76 |
4 |
54 |
1 |
5 |
Determine the linkage relations of these two genes to their centromere(s) and to each other. Specify distances in map units.
Draw a diagram to show the origin of the ascus type with only one single representative (second from right).
Question 4.45
A corn geneticist wants to obtain a corn plant that has the three dominant phenotypes: anthocyanin (A), long tassels (L), and dwarf plant (D). In her collection of pure lines, the only lines that bear these alleles are AA LL dd and aa ll DD. She also has the fully recessive line aa ll dd. She decides to intercross the first two and testcross the resulting hybrid to obtain in the progeny a plant of the desired phenotype (which would have to be Aa Ll Dd in this case). She knows that the three genes are linked in the order written, that the distance between the A/a and the L/l loci is 16 m.u., and that the distance between the L/l and the D/d loci is 24 m.u.
Draw a diagram of the chromosomes of the parents, the hybrid, and the tester.
Draw a diagram of the crossover(s) necessary to produce the desired genotype.
What percentage of the testcross progeny will be of the phenotype that she needs?
What assumptions did you make (if any)?
Question 4.46
In the model plant Arabidopsis thaliana, the following alleles were used in a cross:
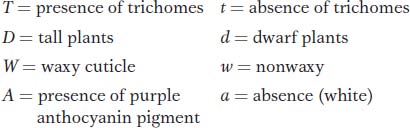
The T/t and D/d loci are linked 26 m.u. apart on chromosome 1, whereas the W/w and A/a loci are linked 8 m.u. apart on chromosome 2.
A pure-
What will be the appearance of the F1?
Sketch the chromosomes 1 and 2 of the parents and the F1, showing the arrangement of the alleles.
If the F1 is testcrossed, what proportion of the progeny will have all four recessive phenotypes?
Question 4.47
In corn, the cross WW ee FF × ww EE ff is made. The three loci are linked as follows:

Assume no interference.
If the F1 is testcrossed, what proportion of progeny will be ww ee ff?
If the F1 is selfed, what proportion of progeny will be ww ee ff?
Question 4.48
The fungal cross + · + × c · m was made, and nonlinear (unordered) tetrads were collected. The results were
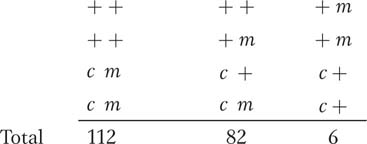
From these results, calculate a simple recombinant frequency.
Compare the Haldane mapping function and the Perkins formula in their conversions of the RF value into a “corrected” map distance.
In the derivation of the Perkins formula, only the possibility of meioses with zero, one, and two crossovers was considered. Could this limit explain any discrepancy in your calculated values? Explain briefly (no calculation needed).
Question 4.49
In mice, the following alleles were used in a cross:
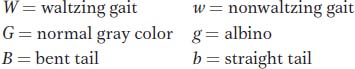
A waltzing gray bent-
|
waltzing |
gray |
bent |
18 |
|
waltzing |
albino |
bent |
21 |
|
nonwaltzing |
gray |
straight |
19 |
|
nonwaltzing |
albino |
straight |
22 |
|
waltzing |
gray |
straight |
4 |
|
waltzing |
albino |
straight |
5 |
|
nonwaltzing |
gray |
bent |
5 |
|
nonwaltzing |
albino |
bent |
6 |
|
Total |
|
|
100 |
What were the genotypes of the two parental mice in the cross?
Draw the chromosomes of the parents.
169
If you deduced linkage, state the map unit value or values and show how they were obtained.
Question 4.50
Consider the Neurospora cross +; + × f; p
It is known that the +/f locus is very close to the centromere on chromosome 7–
parental ditypes showing MI patterns for both loci?
nonparental ditypes showing MI patterns for both loci?
tetratypes showing an MI pattern for +/f and an MI pattern for +/p?
tetratypes showing an MII pattern for +/f and an MI pattern for +/p?
Question 4.51
In a haploid fungus, the genes al-
al-
what proportion of progeny would be prototrophic ++; ++?
Question 4.52
The recessive alleles k (kidney-
|
k |
c |
e |
3 |
|
k |
c |
+ |
876 |
|
k |
+ |
e |
67 |
|
k |
+ |
+ |
49 |
|
+ |
c |
e |
44 |
|
+ |
c |
+ |
58 |
|
+ |
+ |
e |
899 |
|
+ |
+ |
+ |
4 |
|
Total |
|
|
2000 |
Determine the order of the genes and the map distances between them.
Draw the chromosomes of the parents and the F1.
Calculate interference and say what you think of its significance.
Question 4.53
From parents of genotypes A/A · B/B and a/a · b/b, a dihybrid was produced. In a testcross of the dihybrid, the following seven progeny were obtained:
A/a · B/b, a/a · b/b, A/a · B/b, A/a · b/b, a/a · b/b, A/a · B/b, and a/a · B/b
Do these results provide convincing evidence of linkage?
CHALLENGING PROBLEMS
Question 4.54
Use the Haldane map function to calculate the corrected map distance in cases where the measured RF = 5%, 10%, 20%, 30%, and 40%. Sketch a graph of RF against corrected map distance, and use it to answer the question, When should one use a map function?
 Unpacking Problem 55
Unpacking Problem 55
Question 4.55
An individual heterozygous for four genes, A/a · B/b · C/c · D/d, is testcrossed with a/a · b/b · c/c · d/d, and 1000 progeny are classified by the gametic contribution of the heterozygous parent as follows:
|
a · B · C · D |
42 |
|
A · b · c · d |
43 |
|
A · B · C · d |
140 |
|
a · b · c · D |
145 |
|
a · B · c · D |
6 |
|
A · b · C · d |
9 |
|
A · B · c · d |
305 |
|
a · b · C · D |
310 |
Which genes are linked?
If two pure-
breeding lines had been crossed to produce the heterozygous individual, what would their genotypes have been? Draw a linkage map of the linked genes, showing the order and the distances in map units.
Calculate an interference value, if appropriate.
Question 4.56
An autosomal allele N in humans causes abnormalities in nails and patellae (kneecaps) called the nail–
|
nail– |
66% |
|
normal nails and patellae, blood type O |
16% |
|
normal nails and patellae, blood type A |
9% |
|
nail– |
9% |
Fully analyze these data, explaining the relative frequencies of the four phenotypes. (See pages 219-
Question 4.57
Assume that three pairs of alleles are found in Drosophila: x+ and x, y+ and y and z+ and z. As shown by the symbols, each non-
170
On what chromosome of Drosophila are the genes carried?
Draw the relevant chromosomes in the heterozygous female parent, showing the arrangement of the alleles.
Calculate the map distances between the genes and the coefficient of coincidence.
Question 4.58
The five sets of data given in the following table represent the results of testcrosses using parents with the same alleles but in different combinations. Determine the order of genes by inspection—
|
Phenotypes observed in 3- |
Data sets |
||||
|---|---|---|---|---|---|
|
1 |
2 |
3 |
4 |
5 |
|
|
+ + + |
317 |
1 |
30 |
40 |
305 |
|
+ + c |
58 |
4 |
6 |
232 |
0 |
|
+ b + |
10 |
31 |
339 |
84 |
28 |
|
+ b c |
2 |
77 |
137 |
201 |
107 |
|
a + + |
0 |
77 |
142 |
194 |
124 |
|
a + c |
21 |
31 |
291 |
77 |
30 |
|
a b + |
72 |
4 |
3 |
235 |
1 |
|
a b c |
203 |
1 |
34 |
46 |
265 |
Question 4.59
From the phenotype data given in the following table for two 3-
|
1 |
|
2 |
|||
|---|---|---|---|---|---|
|
+ + + |
669 |
b c d |
8 |
||
|
a b + |
139 |
b + + |
441 |
||
|
a + + |
3 |
b + d |
90 |
||
|
+ + c |
121 |
+ cd |
376 |
||
|
+ b c |
2 |
+ + + |
14 |
||
|
a + c |
2280 |
+ + d |
153 |
||
|
a b c |
653 |
+ c + |
65 |
||
|
+ b + |
2215 |
b c + |
141 |
||
Question 4.60
Vulcans have pointed ears (determined by allele P), absent adrenals (determined by A), and a right-
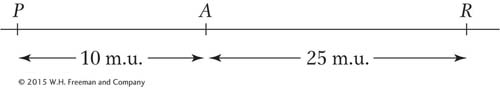
Mr. Spock, first officer of the starship Enterprise, has a Vulcan father and an Earthling mother. If Mr. Spock marries an Earth woman and there is no (genetic) interference, what proportion of their children will have
Vulcan phenotypes for all three characters?
Earth phenotypes for all three characters?
Vulcan ears and heart but Earth adrenals?
Vulcan ears but Earth heart and adrenals?
Question 4.61
In a certain diploid plant, the three loci A, B, and C are linked as follows:
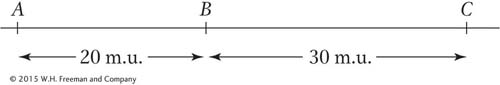
One plant is available to you (call it the parental plant). It has the constitution A b c/a B C.
With the assumption of no interference, if the plant is selfed, what proportion of the progeny will be of the genotype a b c/a b c?
Again, with the assumption of no interference, if the parental plant is crossed with the a b c/a b c plant, what genotypic classes will be found in the progeny? What will be their frequencies if there are 1000 progeny?
Repeat part b, this time assuming 20 percent interference between the regions.
Question 4.62
The following pedigree shows a family with two rare abnormal phenotypes: blue sclerotic (a brittle-
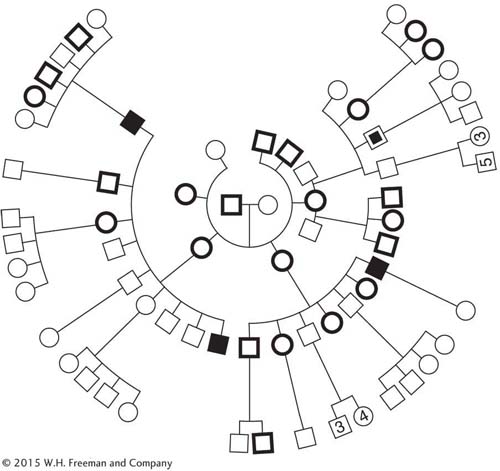
171
What pattern of inheritance is shown by each condition in this pedigree?
Provide the genotypes of as many family members as possible.
Is there evidence of linkage?
Is there evidence of independent assortment?
Can any of the members be judged as recombinants (that is, formed from at least one recombinant gamete)?
Question 4.63
The human genes for color blindness and for hemophilia are both on the X chromosome, and they show a recombinant frequency of about 10 percent. The linkage of a pathological gene to a relatively harmless one can be used for genetic prognosis. Shown here is part of a bigger pedigree. Blackened symbols indicate that the subjects had hemophilia, and crosses indicate color blindness. What information could be given to women III-
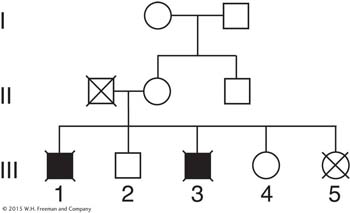
(Problem 63 is adapted from J. F. Crow, Genetics Notes: An Introduction to Genetics. Burgess, 1983.)
Question 4.64
A geneticist mapping the genes A, B, C, D, and E makes two 3-
A/A · B/B · C/C · D/D · E/E × a/a · b/b · C/C · d/d · E/E
The geneticist crosses the F1 with a recessive tester and classifies the progeny by the gametic contribution of the F1:
|
A · B · C · D · E |
316 |
|
a · b · C · d · E |
314 |
|
A · B · C · d · E |
31 |
|
a · b · C · D · E |
39 |
|
A · b · C · d · E |
130 |
|
a · B · C · D · E |
140 |
|
A · b · C · D · E |
17 |
|
a · B · C · d · E |
13 |
|
1000 |
The second cross of pure lines is A/A · B/B · C/C · D/D · E/E × a/a · B/B · c/c · D/D · e/e.
The geneticist crosses the F1 from this cross with a recessive tester and obtains
|
A · B · C · D · E |
243 |
|
a · B · c · D · e |
237 |
|
A · B · c · D · e |
62 |
|
a · B · C · D · E |
58 |
|
A · B · C · D · e |
155 |
|
a · B · c · D · E |
165 |
|
a · B · C · D · e |
46 |
|
A · B · c · D · E |
34 |
|
1000 |
The geneticist also knows that genes D and E assort independently.
Draw a map of these genes, showing distances in map units wherever possible.
Is there any evidence of interference?
Question 4.65
In the plant Arabidopsis, the loci for pod length (L, long; l, short) and fruit hairs (H, hairy; h, smooth) are linked 16 m.u. apart on the same chromosome. The following crosses were made:
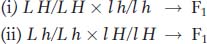
If the F1’s from cross i and cross ii are crossed,
what proportion of the progeny are expected to be l h/l h?
what proportion of the progeny are expected to be L h/l h?
Question 4.66
In corn (Zea mays), the genetic map of part of chromosome 4 is as follows, where w, s, and e represent recessive mutant alleles affecting the color and shape of the pollen:

If the following cross is made
+ + +/+ + + × w s e/w s e
and the F1 is testcrossed with w s e/w s e, and if it is assumed that there is no interference on this region of the chromosome, what proportion of progeny will be of the following genotypes?
+ + +
w s e
+ s e
w + +
+ + e
w s +
w + e
+ s +
Question 4.67
Every Friday night, genetics student Jean Allele, exhausted by her studies, goes to the student union’s bowling lane to relax. But, even there, she is haunted by her genetic studies. The rather modest bowling lane has only four bowling balls: two red and two blue. They are bowled at the pins and are then collected and returned down the chute in random order, coming to rest at the end stop. As the evening passes, Jean notices familiar patterns of the four balls as they come to rest at the stop. Compulsively, she counts the different patterns. What patterns did she see, what were their frequencies, and what is the relevance of this matter to genetics?
172
Question 4.68
In a tetrad analysis, the linkage arrangement of the p and q loci is as follows:
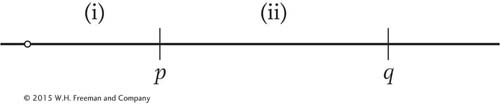
Assume that
in region i, there is no crossover in 88 percent of meioses and there is a single crossover in 12 percent of meioses;
in region ii, there is no crossover in 80 percent of meioses and there is a single crossover in 20 percent of meioses; and
there is no interference (in other words, the situation in one region does not affect what is going on in the other region).
What proportions of tetrads will be of the following types? (a) MIMI, PD; (b) MIMI, NPD; (c) MIMII; (d)MIIMI T; (e) MIIMII, PD; (f) MIIMII, NPD; (g) MIIMII,T. (Note: Here the M pattern written first is the one that pertains to the p locus.) Hint: The easiest way to do this problem is to start by calculating the frequencies of asci with crossovers in both regions, region i, region ii, and neither region. Then determine what MI and MII patterns result.
Question 4.69
For an experiment with haploid yeast, you have two different cultures. Each will grow on minimal medium to which arginine has been added, but neither will grow on minimal medium alone. (Minimal medium is inorganic salts plus sugar.) Using appropriate methods, you induce the two cultures to mate. The diploid cells then divide meiotically and form unordered tetrads. Some of the ascospores will grow on minimal medium. You classify a large number of these tetrads for the phenotypes ARG− (arginine requiring) and ARG+ (arginine independent) and record the following data:
|
Segregation of ARG− : ARG+ |
Frequency (%) |
|---|---|
|
4:0 |
40 |
|
3:1 |
20 |
|
2:2 |
40 |
Using symbols of your own choosing, assign genotypes to the two parental cultures. For each of the three kinds of segregation, assign genotypes to the segregants.
If there is more than one locus governing arginine requirement, are these loci linked?
Question 4.70
An RFLP analysis of two pure lines A/A · B/B and a/a · b/b showed that the former was homozygous for a long RFLP allele (l) and the latter for a short allele (s). The two were crossed to form an F1, which was then backcrossed to the second pure line. A thousand progeny were scored as follows:
|
Aa Bb ss |
9 |
Aa bb ss |
43 |
|
|
Aa Bb ls |
362 |
Aa bb ls |
93 |
|
|
aa bb ls |
11 |
aa Bb ls |
37 |
|
|
aa bb ss |
358 |
aa Bb ss |
87 |
What do these results tell us about linkage?
Draw a map if appropriate.
Incorporate the RFLP fragments into your map.