7.5 The Replisome: A Remarkable Replication Machine
Another hallmark of DNA replication is speed. The time needed for E. coli to replicate its chromosome can be as short as 40 minutes. Therefore, its genome of about 5 million base pairs must be copied at a rate of about 2000 nucleotides per second. From the experiment of Cairns, we know that E. coli uses only two replication forks to copy its entire genome. Thus, each fork must be able to move at a rate of as many as 1000 nucleotides per second. What is remarkable about the entire process of DNA replication is that it does not sacrifice speed for accuracy. How can it maintain both speed and accuracy, given the complexity of the reactions at the replication fork? The answer is that DNA polymerase is part of a large “nucleo-protein” complex that coordinates the activities at the replication fork. This complex, called the replisome, is an example of a “molecular machine.” You will encounter other examples in later chapters. The discovery that most of the major functions of cells—replication, transcription, and translation, for example—are carried out by large multisubunit complexes has changed the way that we think about the cell. To begin to understand why, let’s look at the replisome more closely.
Some of the interacting components of the replisome in E. coli are shown in Figure 7-20. At the replication fork, the catalytic core of DNA pol III is part of a much larger complex, called the pol III holoenzyme, which consists of two catalytic cores and many accessory proteins. One of the catalytic cores handles the synthesis of the leading strand while the other handles lagging-strand synthesis. Some of the accessory proteins (not visible in Figure 7-20) form a connection that bridges the two catalytic cores, thus coordinating the synthesis of the leading and lagging strands. The lagging strand is shown looping around so that the replisome can coordinate the synthesis of both strands and move in the direction of the replication fork. An important accessory protein called the β clamp encircles the DNA like a donut and keeps pol III attached to the DNA molecule. Thus, pol III is transformed from an enzyme that can add only 10 nucleotides before falling off the template (termed a distributive enzyme) into an enzyme that stays at the moving fork and adds tens of thousands of nucleotides (a processive enzyme). In sum, through the action of accessory proteins, the synthesis of both the leading and the lagging strands is rapid and highly coordinated.
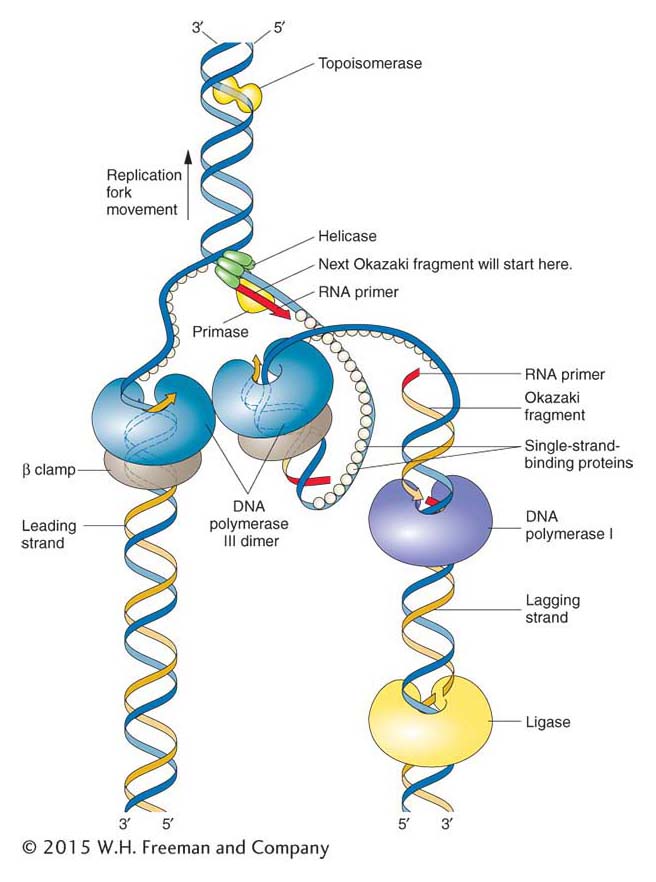
Figure 7-20: Proteins at work at the replication fork
Figure 7-20: The replisome and accessory proteins carry out a number of steps at the replication fork. Topoisomerase and helicase unwind and open the double helix in preparation for DNA replication. When the double helix has been unwound, single-strand-binding proteins prevent the double helix from re-forming. The illustration is a representation of the so-called trombone model (named for its resemblance to a trombone owing to the looping of the lagging strand) showing how the two catalytic cores of the replisome are envisioned to interact to coordinate the numerous events of leading- and lagging-strand replication. ANIMATED ART: Leading- and lagging-strand synthesis
Note that primase, the enzyme that synthesizes the RNA primer, is not touching the clamp protein. Therefore, primase acts as a distributive enzyme—it adds only a few ribonucleotides before dissociating from the template. This mode of action makes sense because the primer need be only long enough to form a suitable duplex starting point for DNA pol III.
Unwinding the double helix
When the double helix was proposed in 1953, a major objection was that the replication of such a structure would require the unwinding of the double helix at the replication fork and the breaking of the hydrogen bonds that hold the strands together. How could DNA be unwound so rapidly and, even if it could, wouldn’t that overwind the DNA behind the fork and make it hopelessly tangled? We now know that the replisome contains two classes of proteins that open the helix and prevent overwinding: they are helicases and topoisomerases, respectively.
Helicases are enzymes that disrupt the hydrogen bonds that hold the two strands of the double helix together. Like the clamp protein, the helicase fits like a donut around the DNA; from this position, it rapidly unzips the double helix ahead of DNA synthesis. The unwound DNA is stabilized by single-strand-binding (SSB) proteins, which bind to single-stranded DNA and prevent the duplex from re-forming.
Circular DNA can be twisted and coiled, much like the extra coils that can be introduced into a twisted rubber band. The unwinding of the replication fork by helicases causes extra twisting at other regions, and supercoils form to release the strain of the extra twisting. Both the twists and the supercoils must be removed to allow replication to continue. This supercoiling can be created or relaxed by enzymes termed topoisomerases, of which an example is DNA gyrase (Figure 7-21). Topoisomerases relax supercoiled DNA by breaking either a single DNA strand or both strands, which allows DNA to rotate into a relaxed molecule. Topoisomerases finish by rejoining the strands of the now relaxed DNA molecule.
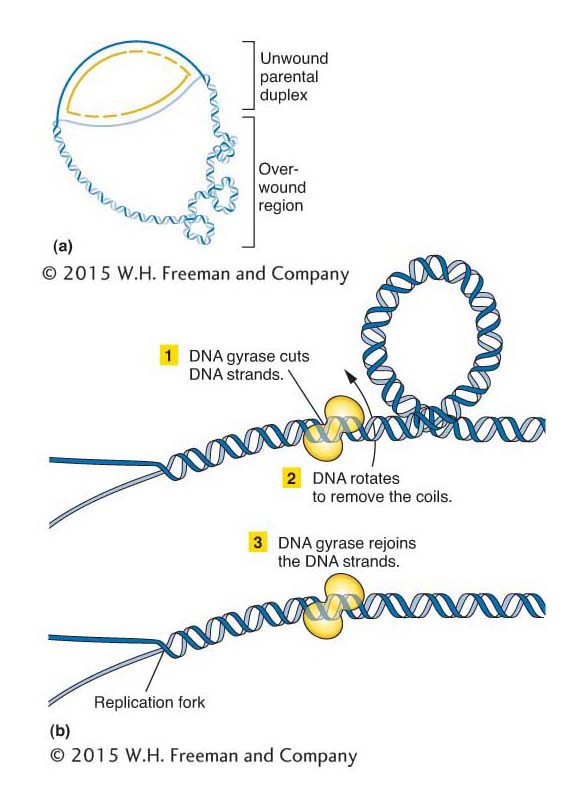
Figure 7-21: DNA gyrase removes extra twists
Figure 7-21: DNA gyrase, a topoisomerase, removes extra twists during replication. (a) Extra-twisted (positively supercoiled) regions accumulate ahead of the fork as the parental strands separate for replication. (b) A topoisomerase such as DNA gyrase removes these regions, by cutting the DNA strands, allowing them to rotate, and then rejoining the strands.
KEY CONCEPT
A molecular machine called the replisome carries out DNA synthesis. It includes two DNA polymerase units to handle synthesis on each strand and coordinates the activity of accessory proteins required for priming, unwinding the double helix, and stabilizing the single strands.
Assembling the replisome: replication initiation
Assembly of the replisome is an orderly process that begins at precise sites on the chromosome (called origins) and takes place only at certain times in the life of the cell. E. coli replication begins from a fixed origin (a locus called oriC) and then proceeds in both directions (with moving forks at both ends, as shown in Figure 7-14) until the forks merge. Figure 7-22 shows the process of replisome assembly. The first step is the binding of a protein called DnaA to a specific 13-base-pair (bp) sequence (called a “DnaA box”) that is repeated five times in oriC. In response to the binding of DnaA, the origin is unwound at a cluster of A and T nucleotides. Recall that AT base pairs are held together with only two hydrogen bonds, whereas GC base pairs are held together with three. Thus, it is easier to separate (melt) the double helix at stretches of DNA that are enriched in A and T bases.
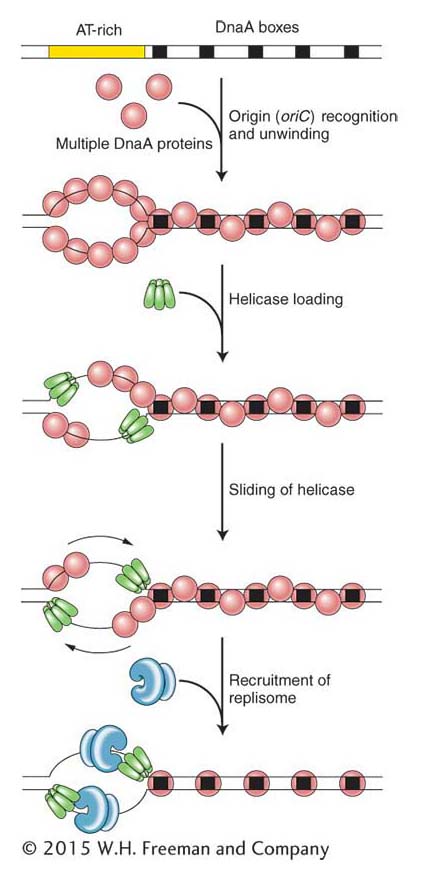
Figure 7-22: Prokaryotic initiation of replication
Figure 7-22: DNA synthesis is initiated at origins of replication in prokaryotes. Proteins bind to the origin (oriC), where they separate the two strands of the double helix and recruit replisome components to the two replication forks.
After unwinding begins, additional DnaA proteins bind to the newly unwound single-stranded regions. With DnaA coating the origin, two helicases (the DnaB protein) now bind and slide in a 5′-to-3′ direction to begin unzipping the helix at the replication fork. Primase and DNA pol III holoenzyme are now recruited to the replication fork by protein–protein interactions, and DNA synthesis begins. You may be wondering why DnaA is not present in Figure 7-20, showing the replisome machine. The answer is that, although it is necessary for the assembly of the replisome, it is not part of the replication machinery. Rather, its job is to bring the replisome to the correct place in the circular chromosome for the initiation of replication.


