Eukaryotic origins of replication
Bacteria such as E. coli usually complete a replication–division cycle in 20 to 40 minutes but, in eukaryotes, the cycle can vary from 1.4 hours in yeast to 24 hours in cultured animal cells and may last from 100 to 200 hours in some cells. Eukaryotes have to solve the problem of coordinating the replication of more than one chromosome.
To understand eukaryotic replication origins, we will first turn our attention to the simple eukaryote yeast. Many eukaryotic proteins having roles at replication origins were first identified in yeast because of the ease of genetic analysis in yeast research (see the yeast Model Organism box in Chapter 12). The origins of replication in yeast are very much like oriC in E. coli. The 100- to 200-bp origins have a conserved DNA sequence that includes an AT-rich region that melts when an initiator protein binds to adjacent binding sites. Unlike prokaryotic chromosomes, each eukaryotic chromosome has many replication origins to replicate the much larger eukaryotic genomes quickly. Approximately 400 replication origins are dispersed throughout the 16 chromosomes of yeast, and there are estimated to be thousands of growing forks in the 23 chromosomes of humans. Thus, in eukaryotes, replication proceeds in both directions from multiple points of origin (Figure 7-23). The double helices that are being produced at each origin of replication elongate and eventually join one another. When replication of the two strands is complete, two identical daughter molecules of DNA result.
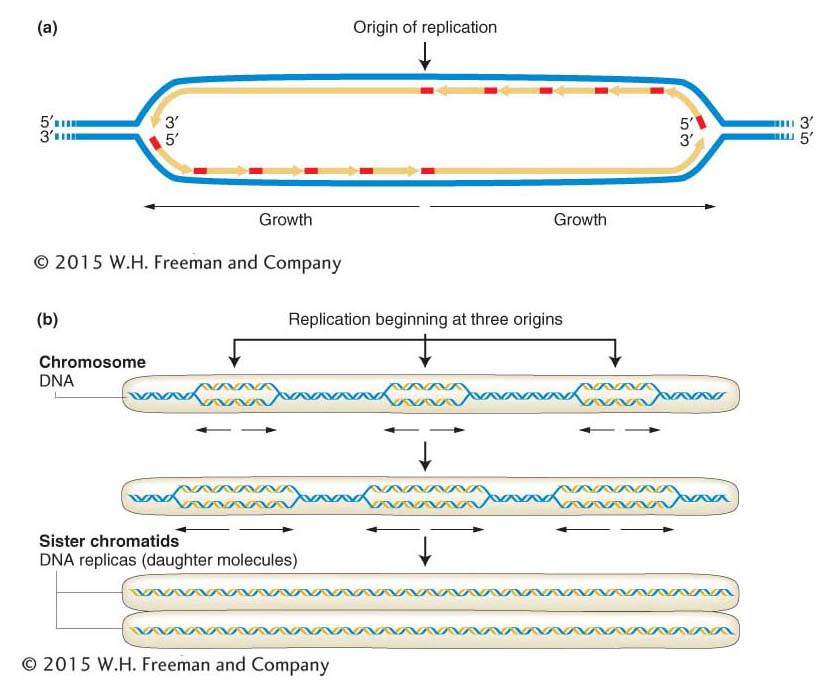
Figure 7-23: DNA replication proceeds in two directions
Figure 7-23: DNA replication proceeds in both directions from an origin of replication. Black arrows indicate the direction of growth of daughter DNA molecules. (a) Starting at the origin, DNA polymerases move outward in both directions. Long yellow arrows represent leading strands and short joined yellow arrows represent lagging strands. (b) How replication proceeds at the chromosome level. Three origins of replication are shown in this example. ANIMATED ART: DNA replication: replication of a chromosome
KEY CONCEPT
Where and when replication takes place are carefully controlled by the ordered assembly of the replisome at a precise site called the origin. Replication proceeds in both directions from a single origin on the circular prokaryotic chromosome. Replication proceeds in both directions from hundreds or thousands of origins on each of the linear eukaryotic chromosomes.
DNA replication and the yeast cell cycle
DNA synthesis takes place in the S (synthesis) phase of the eukaryotic cell cycle (Figure 7-24). How is the onset of DNA synthesis limited to this single stage? In yeast, the method of control is to link replisome assembly to the cell cycle. Figure 7-25 shows the process. In yeast, three proteins are required to begin assembly of the replisome. The origin recognition complex (ORC) first binds to sequences in yeast origins, much as DnaA protein does in E. coli. The presence of ORC at the origin serves to recruit two other proteins, Cdc6 and Cdt1. Both proteins plus ORC then recruit the helicase, called the MCM complex, and the other components of the replisome.
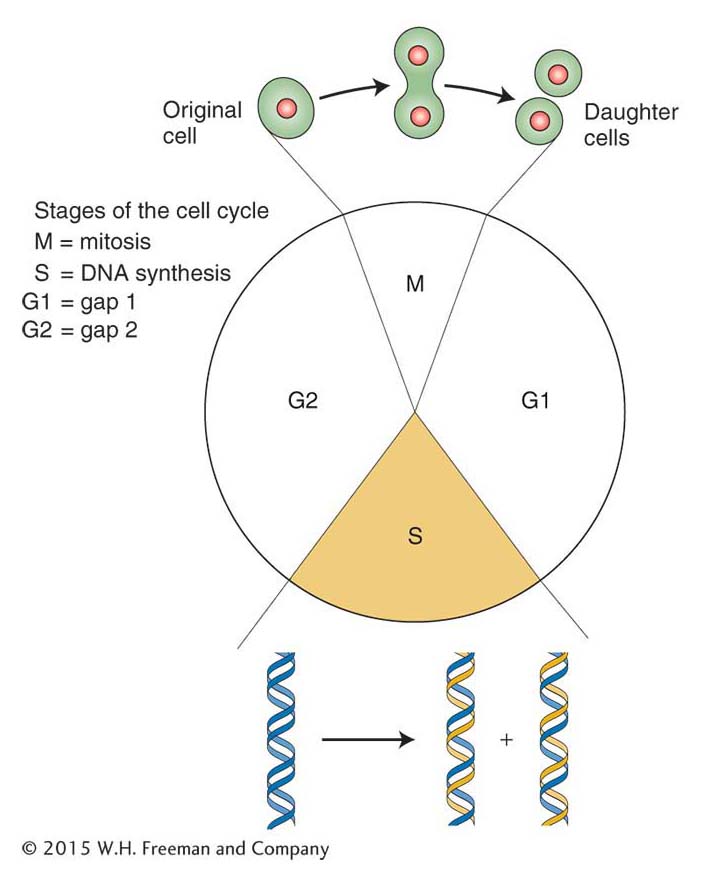
Figure 7-24: Stages of the cell cycle
Figure 7-24: DNA is replicated during the S phase of the cell cycle.
Replication is linked to the cell cycle through the availability of Cdc6 and Cdt1. In yeast, these proteins are synthesized during late mitosis and gap 1 (G1) and are destroyed by proteolysis after synthesis has begun. In this way, the replisome can be assembled only before the S phase. When replication begins, new replisomes cannot form at the origins, because Cdc6 and Cdt1 are degraded during the S phase and are no longer available.
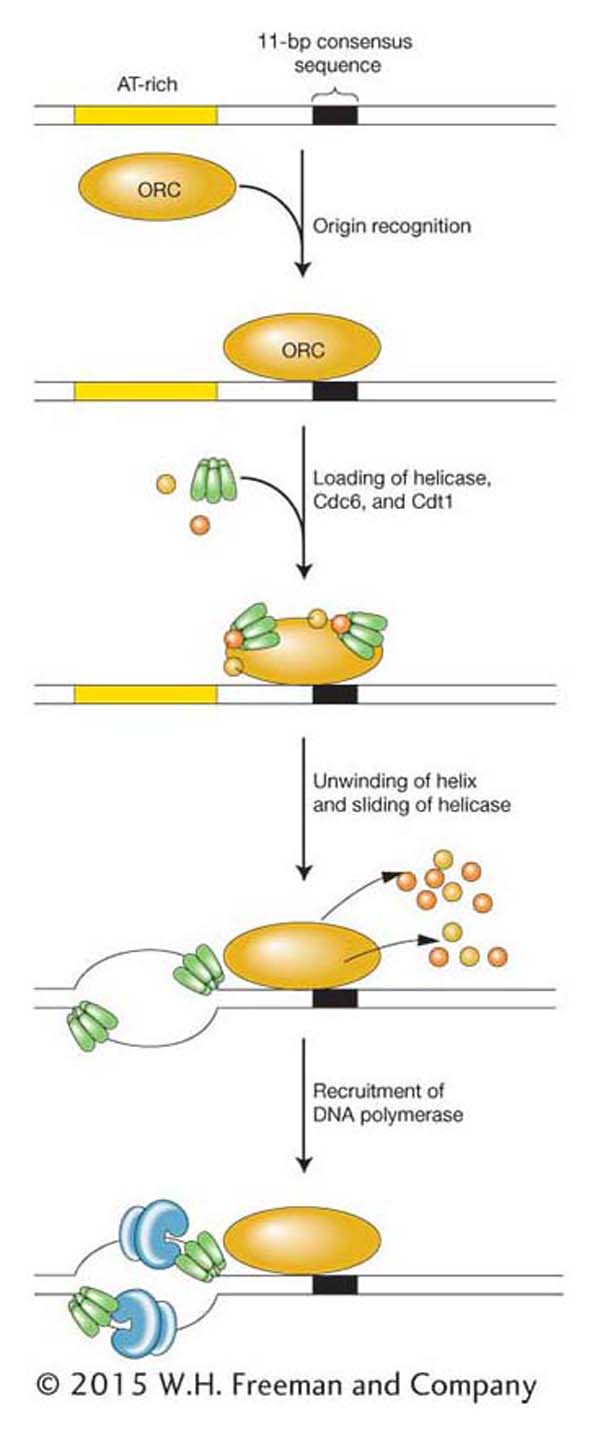
Figure 7-25: Eukaryotic initiation of replication
Figure 7-25: This example from yeast shows the initiation of DNA synthesis at an origin of replication in a eukaryote. As with prokaryotic initiation (see Figure 7-20), proteins of the origin recognition complex (ORC) bind to the origin, where they separate the two strands of the double helix and recruit replisome components at the two replication forks. Replication is linked to the cell cycle through the availability of two proteins: Cdc6 and Cdtl.
Replication origins in higher eukaryotes
As already stated, most of the approximately 400 origins of replication in yeast are composed of similar DNA sequence motifs (100–200 bp in length) that are recognized by the ORC subunits. Interestingly, although all characterized eukaryotes have similar ORC proteins, the origins of replication in higher eukaryotes are much longer, possibly as long as tens of thousands or hundreds of thousands of nucleotides. Significantly, they have limited sequence similarity. Thus, although the yeast ORC recognizes specific DNA sequences in yeast chromosomes, what the related ORCs of higher eukaryotes recognize is not clear at this time, but the feature recognized is probably not a specific DNA sequence. What this uncertainty means in practical terms is that it is much harder to isolate origins from humans and other higher eukaryotes because scientists cannot use an isolated DNA sequence of one human origin, for example, to perform a computer search of the entire human genome sequence to find other origins.
If the ORCs of higher eukaryotes do not interact with a specific sequence scattered throughout the chromosomes, then how do they find the origins of replication? These ORCs are thought to interact indirectly with origins by associating with other protein complexes that are bound to chromosomes. Such a recognition mechanism may have evolved so that higher eukaryotes can regulate the timing of DNA replication during S phase (see Chapter 12 for more about euchromatin and heterochromatin). Gene-rich regions of the chromosome (the euchromatin) have been known for some time to replicate early in S phase, whereas gene-poor regions, including the densely packed heterochromatin, replicate late in S phase. DNA replication could not be timed by region if ORCs were to bind to related sequences scattered throughout the chromosomes. Instead, ORCs may, for example, have a higher affinity for origins in open chromatin and bind to these origins first and then bind to condensed chromatin only after the gene-rich regions have been replicated.
KEY CONCEPT
The yeast origin of replication, like the origin in prokaryotes, contains a conserved DNA sequence that is recognized by the ORC and other proteins needed to assemble the replisome. In contrast, the origins of higher eukaryotes have been difficult to isolate and study because they are long and complex and do not contain a conserved DNA sequence.


