PROBLEMS
WORKING WITH THE FIGURES
Question 9.1
The primary protein structure is shown in Figure 9-3(a). Where in the mRNA (near the 5′ or 3′ end) would a mutation in R2 be encoded?
Question 9.2
In this chapter you were introduced to nonsense suppressor mutations in tRNA genes. However, suppressor mutations also occur in protein-
Question 9.3
Using the quarternary structure of hemoglobin shown in Figure 9-3(d), explain in structural terms how a mutation in the β subunit protein could be suppressed by a mutation in the α subunit gene.
Question 9.4
Transfer RNAs (tRNAs) are examples of RNA molecules that do not encode protein. Based on Figures 9-6 and 9-8, what is the significance of the sequence of tRNA molecules? What do you predict would be the impact on translation of a mutation in one of the bases of one of the stems in the tRNA structure? On the mutant organism?
Question 9.5
The components of prokaryotic and eukaryotic ribosomes are shown in Figure 9-10. Based on this figure, do you think that the large prokaryotic ribosomal RNA (23S rRNA) would be able to substitute for the eukaryotic 28S rRNA? Justify your answer.
346
Question 9.6
Ribosomal RNAs (rRNAs) are another example of a functional RNA molecule. Based on Figure 9-11, what do you think is the significance of the secondary structure of rRNA?
Question 9.7
In Figure 9-12, is the terminal amino acid emerging from the ribosome encoded by the 5′ or 3′ end of the mRNA?
Question 9.8
In Figure 9-12(b), what do you think happens to the tRNA that is released from the E site?
Question 9.9
In Figure 9-17, what do you think happens next to the ribosomal subunits after they are finished translating that mRNA?
Question 9.10
Based on Figure 9-19, can you predict the position of a mutation that would affect the synthesis of one isoform but not the other?
Question 9.11
Based on Figure 9-24, can you predict the position of a mutation that would produce an active protein that was not directed to the correct location?
BASIC PROBLEMS
 Unpacking the Problem
Unpacking the Problem
Question 9.12
Use the codon dictionary in Figure 9-5 to complete the following table. Assume that reading is from left to right and that the columns represent transcriptional and translational alignments.
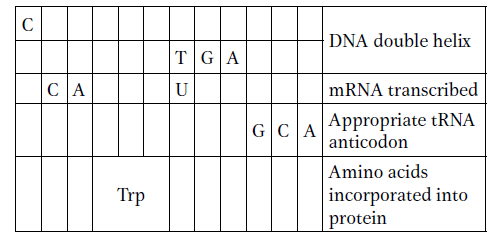
Label the 5′ and 3′ ends of DNA and RNA, as well as the amino and carboxyl ends of the protein.
Question 9.13
Consider the following segment of DNA:
5′ GCTTCCCAA 3′
3′ CGAAGGGTT 5′
Assume that the top strand is the template strand used by RNA polymerase.
Draw the RNA transcribed.
Label its 5′ and 3′ ends.
Draw the corresponding amino acid chain.
Label its amino and carboxyl ends.
Repeat parts a through d, assuming the bottom strand to be the template strand.
Question 9.14
A mutational event inserts an extra nucleotide pair into DNA. Which of the following outcomes do you expect? (1) No protein at all; (2) a protein in which one amino acid is changed; (3) a protein in which three amino acids are changed; (4) a protein in which two amino acids are changed; (5) a protein in which most amino acids after the site of the insertion are changed.
Question 9.15
Before the true nature of the genetic coding process was fully understood, it was proposed that the message might be read in overlapping triplets. For example, the sequence GCAUC might be read as GCA CAU AUC:
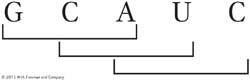
Devise an experimental test of this idea.
Question 9.16
If tRNA is the adaptor for translation, what is the ribosome?
Question 9.17
Which anticodon would you predict for a tRNA species carrying isoleucine? Is there more than one possible answer? If so, state any alternative answers.
Question 9.18
In how many cases in the genetic code would you fail to know the amino acid specified by a codon if you knew only the first two nucleotides of the codon?
In how many cases would you fail to know the first two nucleotides of the codon if you knew which amino acid is specified by it?
Question 9.19
Deduce what the six wild-
Question 9.20
If a polyribonucleotide contains equal amounts of randomly positioned adenine and uracil bases, what proportion of its triplets will encode (a) phenylalanine, (b) isoleucine, (c) leucine, (d) tyrosine?
Question 9.21
You have synthesized three different messenger RNAs with bases incorporated in random sequence in the following ratios: (a) 1 U : 5 C’s, (b) 1 A : 1 C : 4 U’s, (c) 1 A : 1 C : 1 G : 1 U. In a protein-
Question 9.22
In the fungus Neurospora, some mutants were obtained that lacked activity for a certain enzyme. The mutations were found, by mapping, to be in either of two unlinked genes. Provide a possible explanation in reference to quaternary protein structure.
Question 9.23
What is meant by the statement “The genetic code is universal”? What is the significance of this finding?
Question 9.24
The enzyme tryptophan synthetase is produced in two sizes, large and small. Some mutants with no enzyme activity produced exactly the same size enzymes as the wild type. Other mutants with no activity produced just the large enzyme; still others, just the small enzyme.
347
Explain the different types of mutants at the level of protein structure.
Why do you think there were no mutants that produced no enzyme?
Question 9.25
In the Crick-
Question 9.26
A mutant has no activity for the enzyme isocitrate lyase. Does this result prove that the mutation is in the gene encoding isocitrate lyase?
Question 9.27
A certain nonsense suppressor corrects a nongrowing mutant to a state that is near, but not exactly, wild type (it has abnormal growth). Suggest a possible reason why the reversion is not a full correction.
Question 9.28
In bacterial genes, as soon as any partial mRNA transcript is produced by the RNA polymerase system, the ribosome assembles on it and starts translating. Draw a diagram of this process, identifying 5′ and 3′ ends of mRNA, the COOH and NH2 ends of the protein, the RNA polymerase, and at least one ribosome. Why couldn’t this system work in eukaryotes?
Question 9.29
In a haploid, a nonsense suppressor su1 acts on mutation 1 but not on mutation 2 or 3 of gene P. An unlinked nonsense suppressor su2 works on P mutation 2 but not on 1 or 3. Explain this pattern of suppression in regard to the nature of the mutations and the suppressors.
Question 9.30
In vitro translation systems have been developed in which specific RNA molecules can be added to a test tube containing a bacterial cell extract that includes all the components needed for translation (ribosomes, tRNAs, amino acids). If a radioactively labeled amino acid is included, any protein translated from that RNA can be detected and displayed on a gel. If a eukaryotic mRNA is added to the test tube, would radioactive protein be produced? Explain.
Question 9.31
An in vitro translation system contains a eukaryotic cell extract that includes all the components needed for translation (ribosomes, tRNAs, amino acids). If bacterial RNA is added to the test tube, would a protein be produced? If not, why not?
Question 9.32
Would a chimeric translation system containing the large ribosomal subunit from E. coli and the small ribosomal subunit from yeast (a unicellular eukaryote) be able to function in protein synthesis? Explain why or why not.
Question 9.33
Mutations that change a single amino acid in the active site of an enzyme can result in the synthesis of wild-
Question 9.34
What evidence supports the view that ribosomal RNAs are a more important component of the ribosome than the ribosomal proteins?
Question 9.35
Explain why antibiotics, such as erythromycin and Zithromax, that bind the large ribosomal subunit do not harm us.
Question 9.36
Why do multicellular eukaryotes need to have hundreds of kinase-
Question 9.37
Our immune system makes many different proteins that protect us from viral and bacterial infection. Biotechnology companies must produce large quantities of these immune proteins for human testing and eventual sale to the public. To this end, their scientists engineer bacterial or human cell cultures to express these immune proteins. Explain why proteins isolated from bacterial cultures are often inactive, whereas the same proteins isolated from human cell cultures are active (functional).
Question 9.38
Would you expect to find nuclear localization sequences (NLSs) in the proteins that make up prokaryotic and eukaryotic DNA and RNA polymerases? Explain why or why not.
CHALLENGING PROBLEMS
Question 9.39
A single nucleotide addition and a single nucleotide deletion approximately 15 bases apart in the DNA cause a protein change in sequence from
Phe–
to
Phe–
What are the old and new mRNA nucleotide sequences? (Use the codon dictionary in Figure 9-5.)
Which nucleotide has been added? Which has been deleted?
Question 9.40
You are studying an E. coli gene that specifies a protein. A part of its sequence is
–Ala–
You recover a series of mutants for this gene that show no enzymatic activity. By isolating the mutant enzyme products, you find the following sequences:
Mutant 1:
–Ala–
Mutant 2:
–Ala–
Mutant 3:
–Ala–
Mutant 4:
–Ala–
What is the molecular basis for each mutation? What is the DNA sequence that specifies this part of the protein?
Question 9.41
Suppressors of frameshift mutations are now known. Propose a mechanism for their action.
348
Question 9.42
Consider the genes that specify the structure of hemoglobin. Arrange the following events in the most likely sequence in which they would take place.
Anemia is observed.
The shape of the oxygen-
binding site is altered. An incorrect codon is transcribed into hemoglobin mRNA.
The ovum (female gamete) receives a high radiation dose.
An incorrect codon is generated in the DNA of a hemoglobin gene.
A mother (an X-
ray technician) accidentally steps in front of an operating X- ray generator. A child dies.
The oxygen-
transport capacity of the body is severely impaired. The tRNA anticodon that lines up is one of a type that brings an unsuitable amino acid.
Nucleotide-
pair substitution occurs in the DNA of a gene for hemoglobin.
Question 9.43
What structural features are shared by spliceosomes (Figures 8-16 and 8-17) and ribosomes? Why are both structures used to support the RNA World theory?
Question 9.44
A double-
TACATGATCATTTCACGGAATTTCTAGCATGTA
ATGTACTAGTAAAGTGCCTTAAAGATCGTACAT
Which strand of DNA is the template strand, and in which direction is it transcribed?
Label the 5′ and the 3′ ends of each strand.
If an inversion occurs between the second and the third triplets from the left and right ends, respectively, and the same strand of DNA is transcribed, how long will the resultant polypeptide be?
Assume that the original molecule is intact and that the bottom strand is transcribed from left to right. Give the RNA base sequence, and label the 5′ and 3′ ends of the anticodon that inserts the fourth amino acid into the nascent polypeptide. What is this amino acid?
Question 9.45
One of the techniques Khorana used to decipher the genetic code was to synthesize polypeptides in vitro, using synthetic mRNA with various repeating base sequences. For example, (AGA)n, which can be written out as AGAAGAAGAAGAAGA…. Sometimes the resulting polypeptide contained just one amino acid (a homopolymer), and sometimes it contained more than one amino acid (a heteropolymer), depending on the repeating sequence used. Khorana found that sometimes different polypeptides were made from the same synthetic mRNA, suggesting that the initiation of protein synthesis in the system in vitro does not always start at the first nucleotide of the messenger. For example, from (CAA)n, three polypeptides may have been made: aa1 homopolymer (abbreviated aa1-aa1), aa2 homopolymer (aa2-aa2), and aa3 homopolymer (aa3-aa3). These polypeptides probably correspond to the following readings derived by starting at different places in the sequence:
CAA CAA CAA CAA…
ACA ACA ACA ACA…
AAC AAC AAC AAC…
The following table shows the results of Khorana’s experiment.
|
Synthetic mRNA |
Polypeptide(s) synthesized |
|---|---|
|
(UC)n |
(Ser– |
|
(UG)n |
(Val– |
|
(AC)n |
(Thr– |
|
(AG)n |
(Arg– |
|
(UUC)n |
(Ser– |
|
(UUG)n |
(Leu– |
|
(AAG)n |
(Arg– |
|
(CAA)n |
(Thr– |
|
(UAC)n |
(Thr– |
|
(AUC)n |
(Ile– |
|
(GUA)n |
(Ser– |
|
(GAU)n |
(Asp– |
|
(UAUC)n |
(Tyr– |
|
(UUAC)n |
(Leu– |
|
(GAUA)n |
None |
|
(GUAA)n |
None |
|
Note: The order in which the polypeptides or amino acids are listed in the table is not significant except for (UAUC)n and (UUAC)n. |
|
Why do (GUA)n and (GAU)n each encode only two homopolypeptides?
Why do (GAUA)n and (GUAA)n fail to stimulate synthesis?
Using Khorana’s results, assign an amino acid to each triplet in the following list. Remember that there are often several codons for a single amino acid and that the first two letters in a codon are usually the important ones (but that the third letter is occasionally significant). Also keep in mind that some very different-
looking codons sometimes encode the same amino acid. Try to solve this problem without consulting Figure 9-5.
349
|
GUA |
GAU |
UUG |
AAC |
|
GUG |
UUC |
UUA |
GAA |
|
GUU |
AGU |
UAU |
AGA |
|
AUG |
CUU |
AUC |
GAG |
|
UGU |
CUA |
UAC |
CAA |
|
ACA |
UCU |
AAG |
UAG |
|
CAC |
CUC |
ACU |
UGA |
Solving this problem requires both logic and trial and error. Don’t be disheartened: Khorana received a Nobel Prize for doing it. Good luck!
(Data from J. Kuspira and G. W. Walker, Genetics: Questions and Problems. McGraw-
Question 9.46
To construct an “interactome” like the one shown in Figure 9-21, scientists identify all of the protein interactions in a particular tissue or cell type. Comparison of interactomes from human muscle versus human brain tissue reveals very different patterns. If you were the scientist involved in this study, how would you explain these results?
Question 9.47
The genomes of most multicellular eukaryotes encode ~25,000 genes, yet their proteomes contain over 200,000 proteins. Propose two processes that, taken together, account for this discrepancy.
Question 9.48
The exons and introns of a gene are shown below. Alternative splicing of this gene produces three different open reading frames. Predict which exons will form these three mRNAs, and provide justification for your answer. nt = nucleotides.
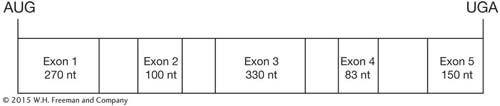
Question 9.49
If life were found on another planet, do you think that it would have the same genetic code? Justify your answer.
Question 9.50
The image on the first page of Chapter 9 is of a ribosome at atomic resolution where the rRNA is blue and the ribosomal proteins are pink. Look at this figure carefully, paying attention to the points of interaction between rRNAs and ribosomal proteins and between ribosomal proteins and tRNAs, mRNAs, and ribosomal factors (EF-
350