9.2 The Genetic Code
The one-
If genes are segments of DNA and if a strand of DNA is just a string of nucleotides, then the sequence of nucleotides must somehow dictate the sequence of amino acids in proteins. How does the DNA sequence dictate the protein sequence? The analogy to a code springs to mind at once. Simple logic tells us that, if the nucleotides are the “letters” in a code, then a combination of letters can form “words” representing different amino acids. First, we must ask how the code is read. Is it overlapping or nonoverlapping? Then we must ask how many letters in the mRNA make up a word, or codon, and which codon or codons represent each amino acid. The cracking of the genetic code is the story told in this section.
325
Overlapping versus nonoverlapping codes
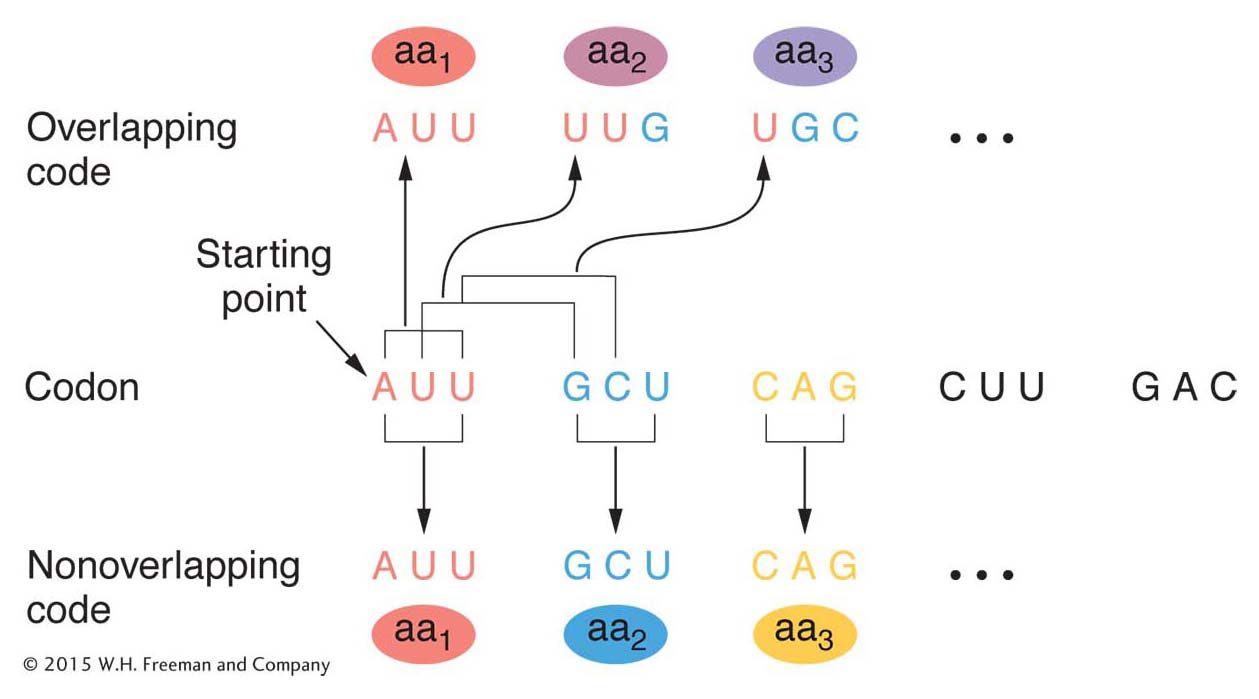
Figure 9-4 shows the difference between an overlapping and a nonoverlapping code. The example shows a three-
By 1961, it was already clear that the genetic code was nonoverlapping. Analyses of mutationally altered proteins showed that only a single amino acid changes at one time in one region of the protein. This result is predicted by a nonoverlapping code. As you can see in Figure 9-4, an overlapping code predicts that a single base change will alter as many as three amino acids at adjacent positions in the protein.
Number of letters in the codon
If an mRNA molecule is read from one end to the other, only one of four different bases, A, U, G, or C, can be found at each position. Thus, if the words encoding amino acids were one letter long, only four words would be possible. This vocabulary cannot be the genetic code because we must have a word for each of the 20 amino acids commonly found in cellular proteins. If the words were two letters long, then 4 × 4 = 16 words would be possible; for example, AU, CU, or CC. This vocabulary is still not large enough.
If the words are three letters long, then 4 × 4 × 4 = 64 words are possible; for example, AUU, GCG, or UGC. This vocabulary provides more than enough words to describe the amino acids. We can conclude that the code word must consist of at least three nucleotides. However, if all words are “triplets,” then the possible words are in considerable excess of the 20 needed to name the common amino acids. We will come back to these excess codons later in the chapter.
Use of suppressors to demonstrate a triplet code
Convincing proof that a codon is, in fact, three letters long (and no more than three) came from beautiful genetic experiments first reported in 1961 by Francis Crick, Sidney Brenner, and their co-
326
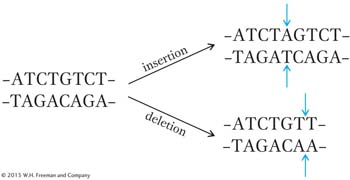
Starting with one particular proflavin-
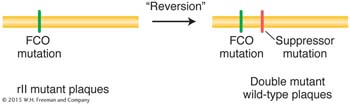
This second mutation “suppressed” mutant expression of the original FCO. Recall from Chapter 6 that a suppressor mutation counteracts or suppresses the effects of another mutation so that the bacterium is more like wild type.
How can we explain these results? If we assume that the gene is read from one end only, then the original addition or deletion induced by proflavin could result in a mutation because it interrupts a normal reading mechanism that establishes the group of bases to be read as words. For example, if each group of three bases on the resulting mRNA makes a word, then the “reading frame” might be established by taking the first three bases from the end as the first word, the next three as the second word, and so forth. In that case, a proflavin-

The insertion suppresses the effect of the deletion by restoring most of the sense of the sentence. By itself, however, the insertion also disrupts the sentence:
THE FAT CAT AAT ETH EBI GRA T
327
If we assume that the FCO mutant is caused by an addition, then the second (suppressor) mutation would have to be a deletion because, as we have seen, only a deletion would restore the reading frame of the resulting message (a second insertion would not correct the frame). In the following diagrams, we use a hypothetical nucleotide chain to represent RNA for simplicity. We also assume that the code words are three letters long and are read in one direction (from left to right in our diagrams).
Wild-
type message CAU CAU CAU CAU CAU
rIIa message: Words after the addition are changed (×) by frameshift mutation (words marked ✓ are unaffected).

rIIarIIb message: Few words are wrong, but reading frame is restored for later words.
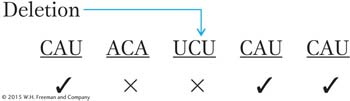
The few wrong words in the suppressed genotype could account for the fact that the “revertants” (suppressed phenotypes) that Crick and his associates recovered did not look exactly like the true wild types in phenotype.
We have assumed here that the original frameshift mutation was an addition, but the explanation works just as well if we assume that the original FCO mutation is a deletion and the suppressor is an addition. You might want to verify it on your own. Very interestingly, combinations of three additions or three deletions have been shown to act together to restore a wild-
Degeneracy of the genetic code
As already stated, with four letters from which to choose at each position, a three-
The reasoning goes like this. If only 20 triplets were used, then the other 44 would be nonsense in that they would not encode any amino acid. In that case, most frameshift mutations could be expected to produce nonsense words, which presumably stop the protein-
328
KEY CONCEPT
The discussion so far demonstrates that1. The linear sequence of nucleotides in a gene determines the linear sequence of amino acids in a protein.
2. The genetic code is nonoverlapping.
3. Three bases encode an amino acid. These triplets are termed codons.
4. The code is read from a fixed starting point and continues to the end of the coding sequence. We know that the code is read sequentially because a single frameshift mutation anywhere in the coding sequence alters the codon alignment for the rest of the sequence.
5. The code is degenerate in that some amino acids are specified by more than one codon.
Cracking the code
The deciphering of the genetic code—
One breakthrough was the discovery of how to make synthetic mRNA. If the nucleotides of RNA are mixed with a special enzyme (polynucleotide phosphorylase), a single-
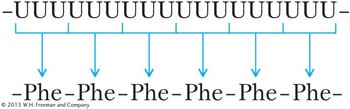
For this discovery, Nirenberg was awarded the Nobel Prize.
Next, mRNAs containing two types of nucleotides in repeating groups were synthesized. For instance, synthetic mRNA having the sequence (AGA)n, which is a long sequence of AGAAGAAGAAGAAGA, was used to stimulate polypeptide synthesis in vitro (in a test tube that also contained a cell extract with all the components necessary for translation). The sequence of the resulting polypeptides was observed from a variety of such tests, with the use of different triplets residing in other synthetic RNAs. From such tests, many code words could be verified. (This kind of experiment is detailed in Problem 44 at the end of this chapter. In solving it, you can put yourself in the place of H. Gobind Khorana, who received a Nobel Prize for directing the experiments.)
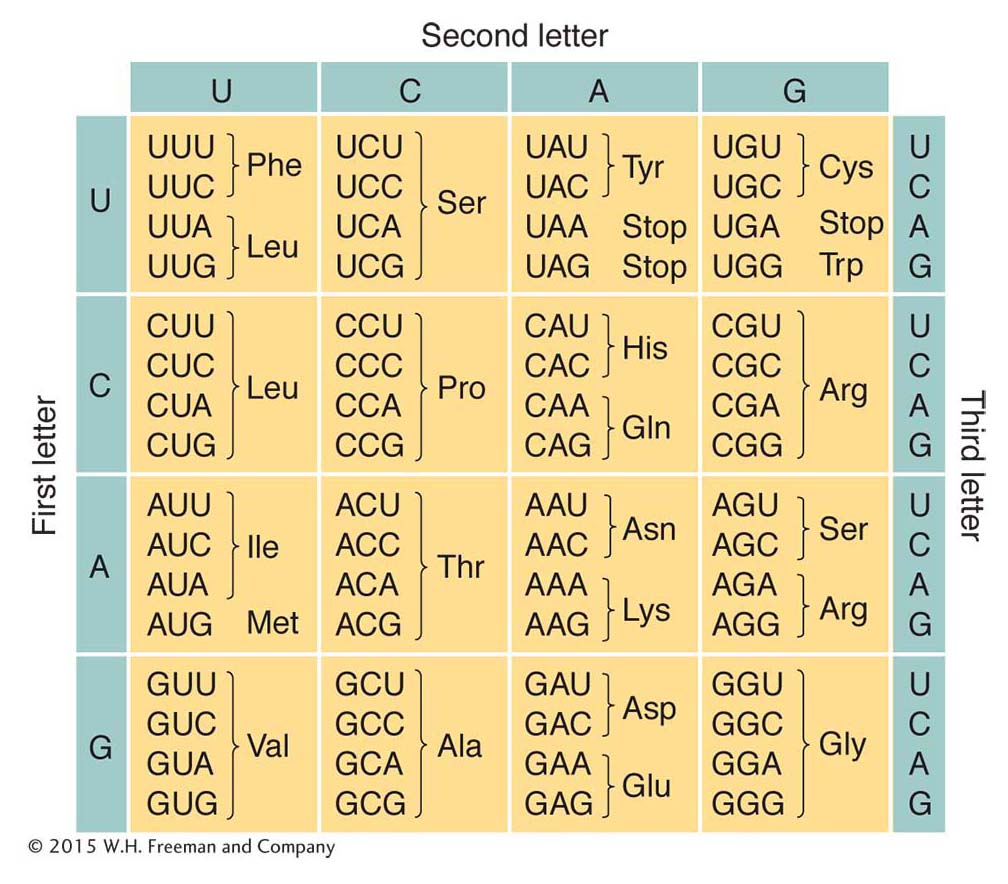
Additional experimental approaches led to the assignment of each amino acid to one or more codons. Recall that the code was proposed to be degenerate, meaning that some amino acids had more than one codon assignment. This degeneracy can be seen clearly in Figure 9-5, which gives the codons and the amino acids that they specify. Virtually all organisms on Earth use this same genetic code. (There are just a few exceptions in which a small number of the codons have different meanings—
329
Stop codons
You may have noticed in Figure 9-5 that some codons do not specify an amino acid at all. These codons are stop, or termination, codons. They can be regarded as being similar to periods or commas punctuating the message encoded in the DNA.
One of the first indications of the existence of stop codons came in 1965 from Brenner’s work with the T4 phage. Brenner analyzed certain mutations (m1–m6) in a single gene that controls the head protein of the phage. He found that the head protein of each mutant was a shorter polypeptide chain than that of the wild type. Brenner examined the ends of the shortened proteins and compared them with the wild-
UAG was the first stop codon deciphered; it is called the amber codon (amber is the English translation of the last name of the codon’s discoverer, Bernstein). Mutants that are defective owing to the presence of an abnormal amber codon are called amber mutants. Two other stop codons are UGA and UAA. Analogously to the amber codon, and continuing the theme of naming for colors and gems, UGA is called the opal codon and UAA is called the ochre codon. Mutants that are defective because they contain abnormal opal or ochre codons are called opal and ochre mutants, respectively. Stop codons are often called nonsense codons because they designate no amino acid.
In addition to a shorter head protein, Brenner’s phage mutants had another interesting feature in common: the presence of a suppressor mutation (su−) in the host chromosome would cause the phage to develop a head protein of normal (wild-