PROBLEMS
WORKING WITH THE FIGURES
Question 12.1
In Figure 12-4, certain mutations decrease the relative transcription rate of the β-globin gene. Where are these mutations located, and how do they exert their effects on transcription?
Question 12.2
Based on the information in Figure 12-6, how does Gal4 regulate four different GAL genes at the same time? Contrast this mechanism with how the Lac repressor controls the expression of three genes.
Question 12.3
In any experiment, controls are essential in order to determine the specific effect of changing some parameter. In Figure 12-7, which constructs are the “controls” that serve to establish the principle that activation domains are modular and interchangeable?
Question 12.4
Contrast the role of the MCM1 protein in different yeast cell types shown in Figure 12-10. How are the a-specific genes controlled differently in different cell types?
Question 12.5
In Figure 12-11b, in what chromosomal region are you likely to find the most H1 histone protein?
Question 12.6
What is the conceptual connection between Figures 12-12 and 12-19?
Question 12.7
In Figure 12-19, where is the TATA box located before the enhanceosome forms at the top of the figure?
Question 12.8
Let’s say that you have incredible skill and can isolate the white and red patches of tissue from the Drosophila eyes shown in Figure 12-24 in order to isolate mRNA from each tissue preparation. Using your knowledge of DNA techniques from Chapter 10, design an experiment that would allow you to determine whether RNA is transcribed from the white gene in the red tissue or the white tissue or both. If you need it, you have access to radioactive white-
Question 12.9
In Figure 12-26, provide a biochemical mechanism for why HP-
Question 12.10
In reference to Figure 12-28, draw the outcome if there is a mutation in gene B.
BASIC PROBLEMS
Question 12.11
What analogies can you draw between transcriptional trans-
Question 12.12
Contrast how the ground states of genes within DNA in bacteria and eukaryotes differ with respect to gene activation (see Figure 12-2).
Question 12.13
Predict and explain the effect on GAL1 transcription, in the presence of galactose alone, of the following mutations:
Deletion of one Gal4-
binding site in the GAL1 UAS element. Deletion of all four Gal4-
binding sites in the GAL1 UAS element. Deletion of the Mig1-
binding site upstream of GAL1. Deletion of the Gal4 activation domain.
Deletion of the GAL80 gene.
Deletion of the GAL1 promoter.
Deletion of the GAL3 gene.
467
Question 12.14
How is the activation of the GAL1 gene prevented in the presence of galactose and glucose?
Question 12.15
What are the roles of histone deacetylation and histone acetylation in gene regulation, respectively?
Question 12.16
An α strain of yeast that cannot switch mating type is isolated. What mutations might it carry that would explain this phenotype?
Question 12.17
What genes are regulated by the α1 and α2 proteins in an α cell?
Question 12.18
What are Sir proteins? How do mutations in SIR genes affect the expression of mating-
Question 12.19
What is meant by the term epigenetic inheritance? What are two examples of such inheritance?
Question 12.20
What is an enhanceosome? Why could a mutation in any one of the enhanceosome proteins severely reduce the transcription rate?
Question 12.21
Usually, the deletion of a gene results in a recessive mutation. Explain how the deletion of an imprinted gene can be a dominant mutation.
Question 12.22
What has to happen for the expression of two different genes on two different chromosomes to be regulated by the same miRNA?
Question 12.23
What mechanisms are thought to be responsible for the inheritance of epigenetic information?
Question 12.24
What is the fundamental difference in how bacterial and eukaryotic genes are regulated?
Question 12.25
Why is it said that transcriptional regulation in eukaryotes is characterized by combinatorial interactions?
Question 12.26
Which of the following statements about histones is true?
They are proteins whose sequence is highly conserved in all eukaryotes.
They are the building blocks of nucleosomes.
a and b are correct.
None of the above are correct.
Question 12.27
Nucleosomes are
composed of DNA and protein.
necessary to condense DNA/chromosomes.
essential for the correct regulation of eukaryotic gene expression.
All of the above are true.
Question 12.28
The regions of chromosomes that form heterochromatin
contain highly expressed genes.
contain few genes.
are associated with the nuclear envelope.
are abundant in prokaryotes.
Question 12.29
Dosage compensation is necessary because
some regions of the genome contain more genes than others.
genes near heterochromatin tend to be silenced.
enhancers can activate transcription whether they are upstream or downstream of a gene.
genes on the X chromosome have twice as many copies in females as in males.
Question 12.30
Which of the following is true of chromatin in prokaryotes?
Bacterial chromosomes are not organized into chromatin.
It is found in the nucleus.
It is found in the cytoplasm.
It is very similar to chromatin in eukaryotes.
Question 12.31
Which of the following statements is/are true of the variegated eye color phenotype in Figure 12-23?
The white gene is active in the white sectors and inactive in the red sectors.
The white gene is inactive in the white sectors and active in the red sectors.
The white gene is inactive in the white sectors due to the spread of heterochromatin.
b and c are both true.
Question 12.32
Which of the following are epigenetic marks?
Methylated basic amino acids in histone tails
Methylated cytosine residues in DNA
Acetylated amino acids in histone tails
All of the above are epigenetic marks.
CHALLENGING PROBLEMS
Question 12.33
The transcription of a gene called YFG (your favorite gene) is activated when three transcription factors (TFA, TFB, TFC) interact to recruit the co-
Question 12.34
A single mutation in one of the transcription factors in Problem 33 results in a drastic reduction in YFG transcription. Diagram what this mutant interaction might look like.
Question 12.35
Diagram the effect of a mutation in the binding site for one of the transcription factors in Problem 33.
Question 12.36
Null alleles (mutant genes) produce no protein product. This is a genetic change. However, epigenetically silenced genes also produce no protein product. How does one determine experimentally whether a gene has been silenced by mutation or has been silenced epigenetically?
Question 12.37
What are epigenetic marks? Which are associated with heterochromatin? How are epigenetic marks thought to be responsible for determining chromatin structure?
468
Question 12.38
You receive four strains of yeast in the mail, and the accompanying instructions state that each strain contains a single copy of transgene A. You grow the four strains and determine that only three strains express the protein product of transgene A. Further analysis reveals that transgene A is located at a different position in the yeast genome in each of the four strains. Provide a hypothesis to explain this result.
Question 12.39
You wish to find the cis-
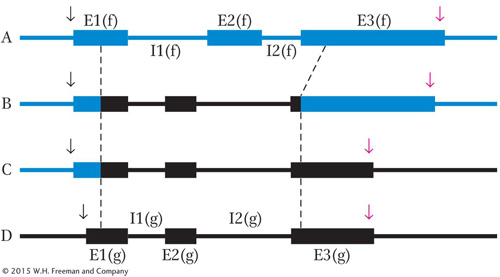
You introduce all four of these clones simultaneously into tissue-
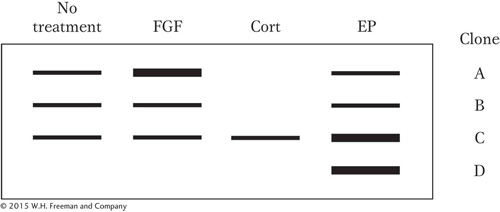
The levels of transcripts produced from the introduced genes in response to various treatments are shown; the intensity of these bands is proportional to the amount of transcript made from a particular clone. (The failure of a band to appear indicates that the level of transcript is undetectable.)
Where is the DNA element that permits activation by FGF?
Where is the DNA element that permits repression by Cort?
Where is the DNA element that permits induction by EP? Explain your answer.
Question 12.40
Using the experimental system shown in Figure 12-26, a geneticist is able to insert a barrier insulator between the white gene and the heterochromatin. The eye phenotype of the transgenic Drosophila would most likely be
all white because the barrier element will prevent the spread of heterochromatin.
all red because the barrier element will prevent the spread of heterochromatin.
still variegated because barrier elements don’t stop the spread of heterochromatin.
still variegated because barrier elements don’t work in Drosophila.
Question 12.41
Which of the following is an example of post-
The amount of RNA transcribed from a gene is reduced because the DNA is methylated.
The amount of RNA is reduced because it is rapidly degraded.
The amount of protein made is reduced by action of an miRNA.
b and c are both correct.