12.4 Activation of Genes in a Chromatin Environment
As you have seen in this chapter, the transcription of eukaryotic genes has to be turned on and off during the lifetime of an organism. To understand how eukaryotes regulate genes during their lifetime, it is necessary to see how chromatin changes during transcriptional activation. In addition, the development of a complex organism requires that transcription levels be regulated over a wide range of activities. Think of a regulation mechanism as more like a rheostat that controls sound from an iPod than an on-or-off switch: rather than a gene producing either many or no proteins, it may produce a number anywhere in between depending on the transcription level. In eukaryotes, transcription levels are made finely adjustable in a chromatin environment by clustering binding sites into enhancers. Several different transcription factors or several molecules of the same transcription factor may bind to adjacent sites. The binding of these factors to sites that are the correct distance apart leads to an amplified, or superadditive, effect on activating transcription. When an effect is greater than additive, it is said to be synergistic.
The binding of multiple regulatory proteins to the multiple binding sites in an enhancer can catalyze the formation of an enhanceosome, a large protein complex that acts synergistically to activate transcription. In Figure 12-18 you can see how architectural proteins bend the DNA to promote cooperative interactions between the other DNA-binding proteins. In this mode of enhanceosome action, transcription is activated to very high levels only when all the proteins are present and touching one another in just the right way.
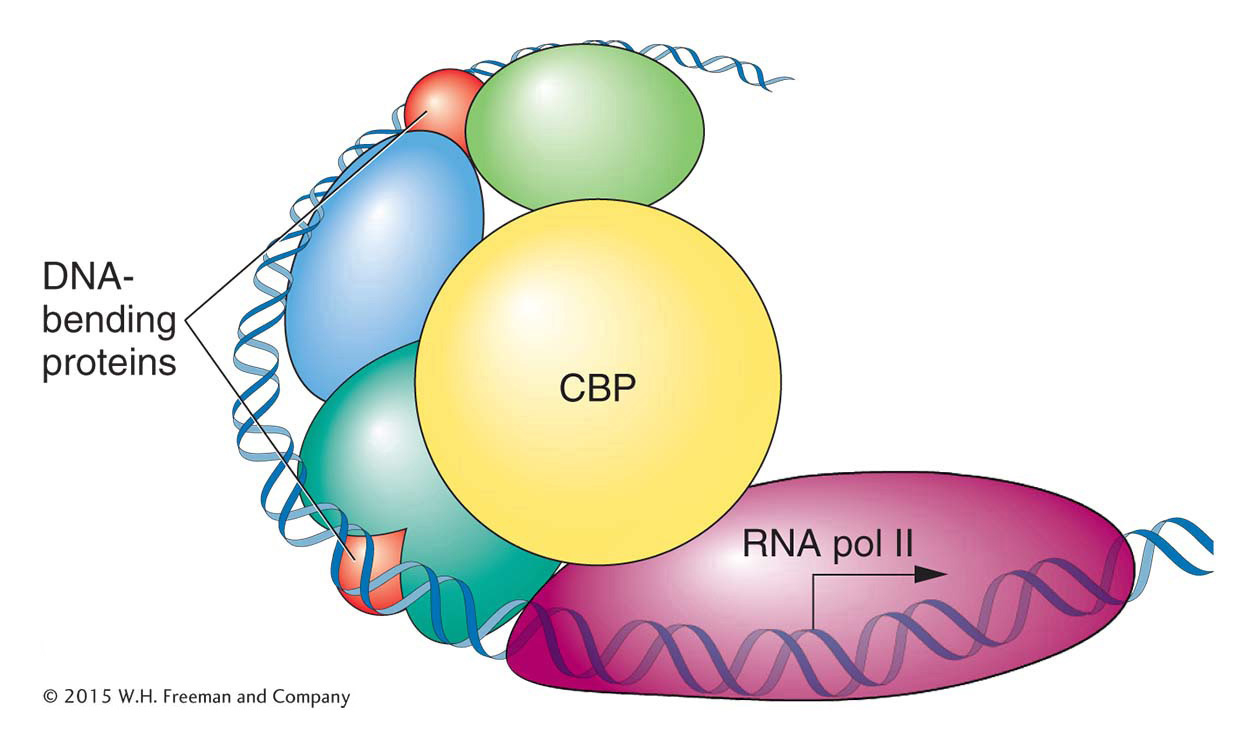
Figure 12-18: Enhanceosomes help recruit the transcriptional machinery
Figure 12-18: The β-interferon enhanceosome. In this case, the transcription factors recruit a co-activator (CBP), which binds both to the transcription factors and to RNA polymerase II, initiating transcription.
To better understand what an enhanceosome is and how it acts synergistically, let’s look at a specific example.
The β-interferon enhanceosome
The human β-interferon gene, which encodes the antiviral protein interferon, is one of the best-characterized genes in eukaryotes. It is normally switched off but is activated to very high levels of transcription on viral infection. The key to the activation of this gene is the assembly of transcription factors into an enhanceosome about 100 bp upstream of the TATA box and transcription start site. The regulatory proteins of the β-interferon enhanceosome all bind to the same face of the DNA double helix. Binding to the other side of the helix are several architectural proteins that bend the DNA and allow the different regulatory proteins to touch one another and form an activated complex. When all of the regulatory proteins are bound and interacting correctly, they form a “landing pad,” a high-affinity binding site for the protein CBP, a co-activator protein that also recruits the transcriptional machinery. The large CBP protein also contains an intrinsic histone acetylase activity that modifies nucleosomes and facilitates high levels of transcription.
Although the β-interferon promoter is shown without nucleosomes in Figure 12-18, the enhanceosome is actually surrounded by two nucleosomes, called nuc 1 and nuc 2 in Figure 12-19. One of them, nuc 2, is strategically positioned over the TATA box and transcription start site. GCN5, another co-activator, binds and acetylates the two nucleosomes. After acetylation, the activating transcription factors recruit the co-activator CBP, the RNA pol II holoenzyme, and the SWI–SNF chromatin-remodeling complex. SWI-SNF is then positioned to nudge the nucleosome 37 bp off the TATA box, making the TATA box accessible to the TATA-binding protein and allowing transcription to be initiated.
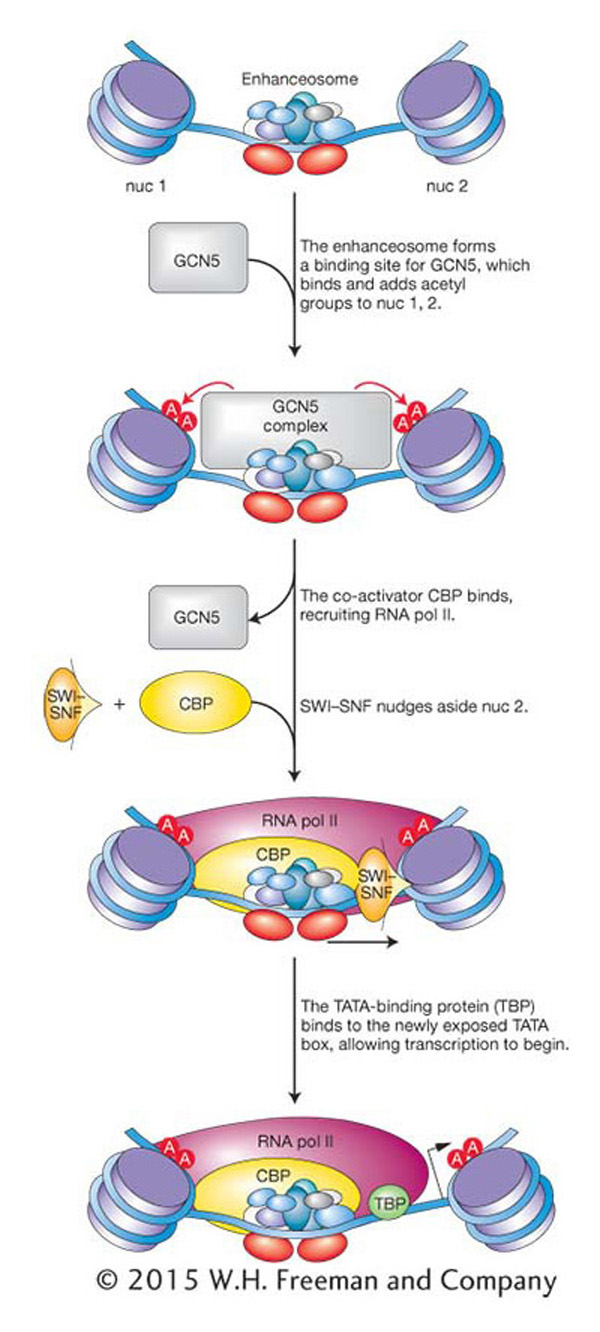
Figure 12-19: Enhanceosomes recruit chromatin remodelers
Figure 12-19: The β-interferon enhanceosome acts to move nucleosomes by recruiting the SWI-SNF complex.
Cooperative interactions help to explain several perplexing observations about enhancers. For example, they explain why mutating any one transcription factor or binding site dramatically reduces enhancer activity. They also explain why the distance between binding sites within the enhancer is such a critical feature. Furthermore, enhancers do not have to be close to the start site of transcription, as is the example shown in Figure 12-19. One characteristic of enhancers is that they can activate transcription when they are located at great distances from the promoter (>50 kb), either upstream or downstream from a gene or even in an intron.
Enhancer-blocking insulators
A regulatory element, such as an enhancer, that can act over tens of thousands of base pairs could interfere with the regulation of nearby genes. To prevent such promiscuous activation, regulatory elements called enhancer-blocking insulators have evolved. When positioned between an enhancer and a promoter, enhancer-blocking insulators prevent the enhancer from activating transcription at that promoter. Such insulators have no effect on the activation of other promoters that are not separated from their enhancers by the insulator (Figure 12-20). Several models have been proposed to explain how an insulator could block enhancer activity only when placed between an enhancer and a promoter. Many of the models, like the one shown in Figure 12-21, propose that the DNA is organized into loops containing active genes. According to this model, insulators act by moving a promoter into a new loop, where it is shielded from the enhancer.
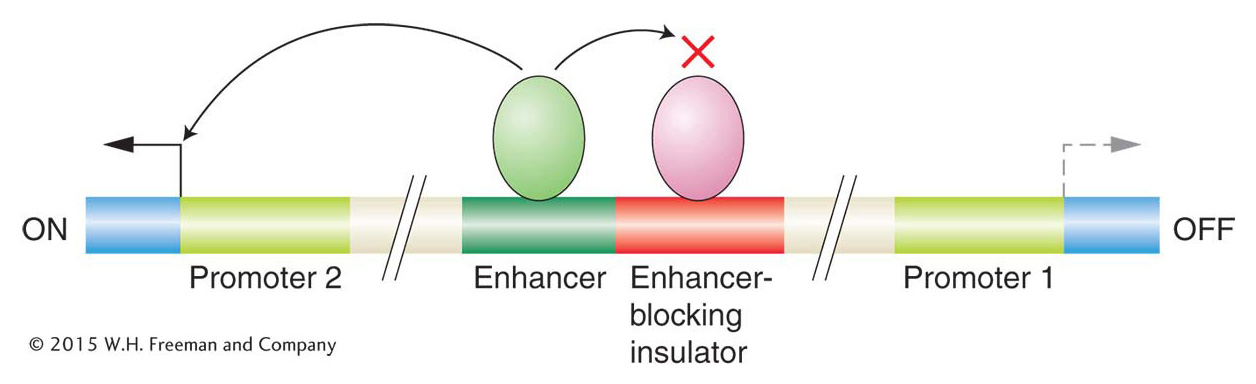
Figure 12-20: Enhancer-blocking insulators prevent enhancer activation
Figure 12-20: Enhancer-blocking insulators prevent gene activation when placed between an enhancer and a promoter.
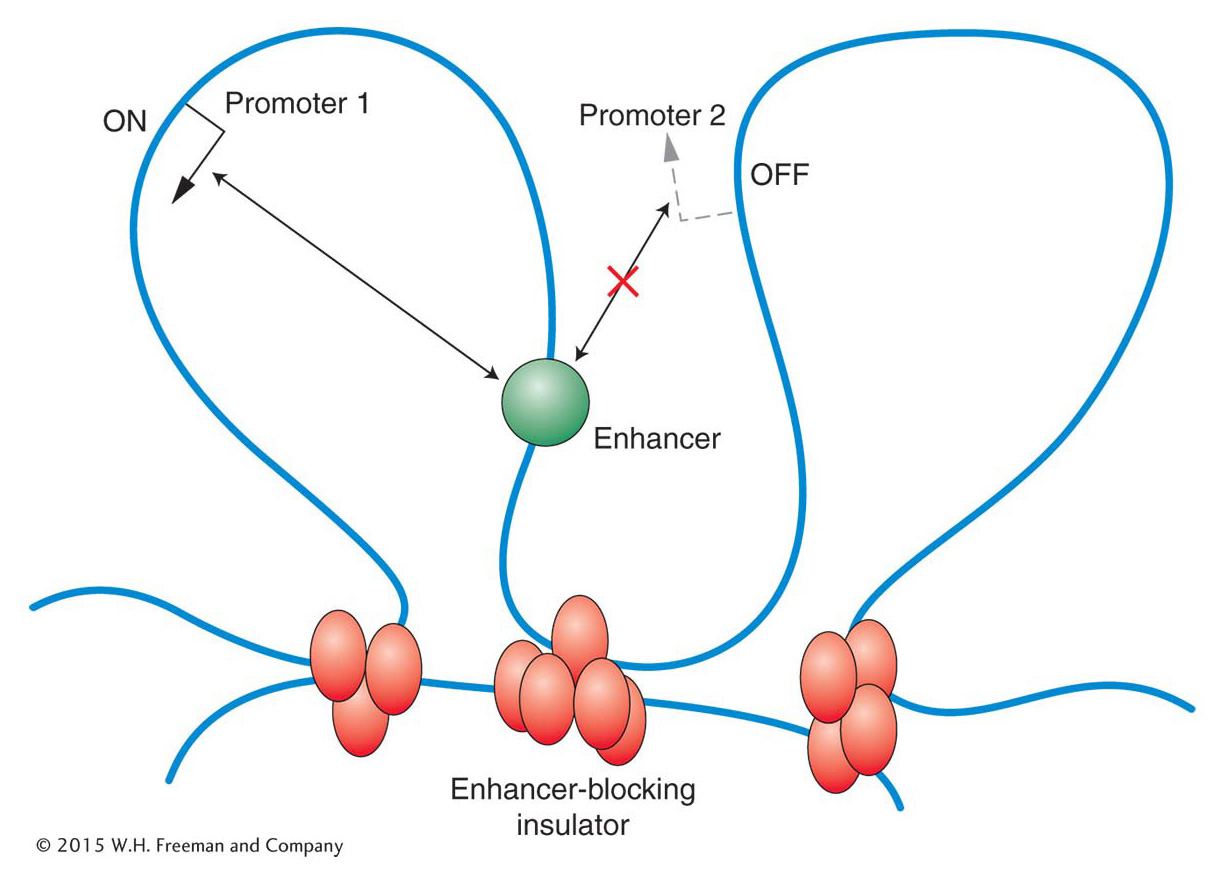
Figure 12-21: Model for how enhancer-blocking insulators might work
Figure 12-21: One proposal is that enhancer-blocking insulators create new loops that physically separate a promoter from its enhancer.
As you will see later in the chapter, enhancer-blocking insulators are a fundamental component of a phenomenon called genomic imprinting.
KEY CONCEPT
Eukaryotic enhancers can act at great distances to modulate the activity of the transcriptional apparatus. Enhancers contain binding sites for many transcription factors, which bind and interact cooperatively. These interactions result in a variety of responses, including the recruitment of additional co-activators and the remodeling of chromatin.



