12.7 Post-Transcriptional Gene Repression by miRNAs
Xist is one example of the rapidly growing class of functional RNAs (see Chapter 8). Functional RNAs do not encode proteins: rather, they perform a variety of tasks that exploit the complementarity of RNA and RNA and of RNA and DNA. The functional RNAs discussed here contain specific sequences that direct proteins or protein complexes to places in the cell where their services are needed. For example, Xist acts to direct proteins involved in heterochromatin formation to one of the two X chromosomes.
464
Two types of small functional RNAs were introduced in Chapter 8: siRNAs and miRNAs. In this section we explore how miRNAs assist in the regulation of eukaryotic gene expression. The function of siRNAs will be considered further in Chapter 15.
Recall from Chapter 8 that miRNAs are synthesized by RNA pol II as longer RNAs that are processed into the smaller (~22 nt) biologically active miRNAs (see Figure 8-
In Chapter 8 we saw how the active single-
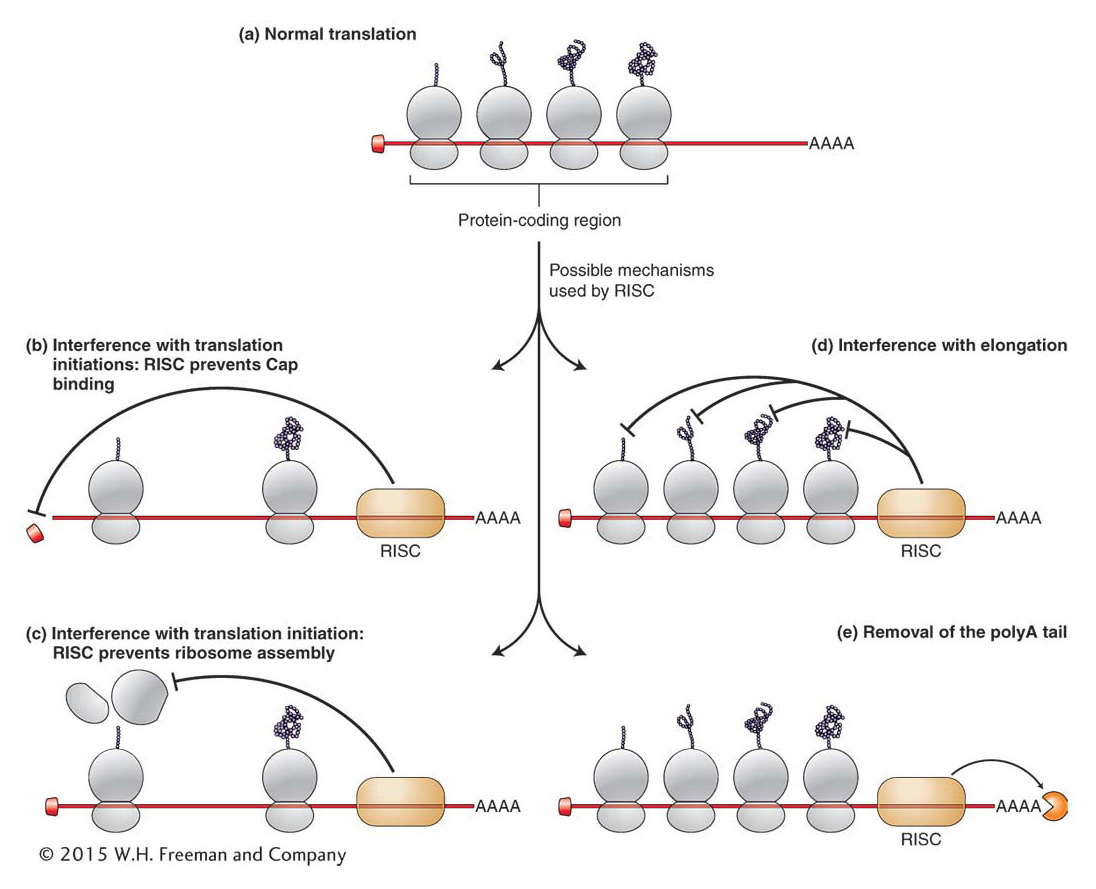
465
Although miRNAs were discovered almost 20 years ago, scientists are only beginning to decipher the extent and complexity of miRNA control of eukaryotic gene expression. Consider that in mammals, sequences complementary to the seed regions of miRNAs are found in the 3′ UTR of several hundred genes. Furthermore, the 3′UTRs of some genes contain sequences complementary to several miRNAs, while many miRNAs contain sequences complementary to the 3′ UTRs of several genes. Thus, one gene can potentially be repressed by several miRNAs (either individually or in combination) and one miRNA can potentially repress the translation of several genes. In Chapter 11 you saw that the organization of bacterial genes into operons permitted the coordinate regulation of genes that contributed to a single trait, such as the ability to utilize the sugar lactose. Given that most eukaryotic genes are not organized into operons, it has been suggested that post-